Applied Study Conditions
R. Noah Padgett
2022-01-31
Last updated: 2022-02-16
Checks: 5 1
Knit directory: Padgett-Dissertation/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210401) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the results is critical for reproducibility. To start using Git, open the Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full reproducibility benefits of using workflowr, please see ?wflow_start.
# Load packages & utility functions
source("code/load_packages.R")
source("code/load_utility_functions.R")
# environment options
options(scipen = 999, digits=3)library(readxl)
mydata <- read_excel("data/temp_data_extracted.xlsx") %>%
filter(is.na(EXCLUSION_FLAG)==T)Journal Information
mydata %>%
select(Journal) %>%
table().
Assessment EJPA JID JPA PA PID
51 10 24 25 12 22 mydata %>%
select(`Analysis (CFA, EFA, IRT, SEM, etc.)`) %>%
table().
CFA CFA/IRT Correlation
70 8 2
EFA EFA/CFA ESEM/CFA
8 5 1
IRT LCA mediation
9 1 1
Moken Scale random-effects models Rasch
1 1 4
regression SEM
3 11 Data Conditions
f <- function(x){
c(mean(x, na.rm=T), sd(x, na.rm=T), quantile(x, c(0, 0.25, 0.5, 0.75, 1), na.rm=T))
}
sum_dat <- mydata %>%
summarise(
SS = f(`Sample Size`),
NLV = f(`Number of Latent Variables`),
NOV = f(`Number of Observed Variables`),
Min_Ind = f(`Minimum Number of Indicators per Factor`),
Avg_Ind = f(`Average Number of Indicators per Factor`),
Max_Ind = f(`Maximum Number of Indicators per Factor`),
Min_FL = f(abs(`Minimum Factor Loading`)),
Avg_FL = f(`Average Factor Loading`),
Max_FL = f(`Maximum Factor Loading`),
Min_Rel = f(`Min Reliability`),
Avg_Rel = f(`Avg Reliability`),
Max_Rel = f(`Max Reliability`)
) %>% as.data.frame()
rownames(sum_dat) <- c("avg", "sd", "min", "q1", "median", "q3", "max")
sum_dat <- t(sum_dat)
kable(sum_dat, format="html", digits=2) %>%
kable_styling(full_width = T)| avg | sd | min | q1 | median | q3 | max | |
|---|---|---|---|---|---|---|---|
| SS | 4217.78 | 23241.20 | 36.00 | 318.00 | 603.00 | 1451.00 | 263683.00 |
| NLV | 5.46 | 6.93 | 1.00 | 2.00 | 4.00 | 6.00 | 60.00 |
| NOV | 50.12 | 81.58 | 5.00 | 12.00 | 22.50 | 41.75 | 442.00 |
| Min_Ind | 6.86 | 6.40 | 1.00 | 4.00 | 5.00 | 8.00 | 40.00 |
| Avg_Ind | 8.95 | 7.00 | 2.50 | 4.66 | 6.22 | 10.93 | 40.00 |
| Max_Ind | 12.93 | 15.20 | 3.00 | 5.00 | 8.00 | 14.00 | 135.00 |
| Min_FL | 0.41 | 0.20 | 0.00 | 0.28 | 0.41 | 0.51 | 0.86 |
| Avg_FL | 0.64 | 0.13 | 0.23 | 0.56 | 0.65 | 0.72 | 0.93 |
| Max_FL | 0.85 | 0.10 | 0.49 | 0.80 | 0.87 | 0.92 | 1.08 |
| Min_Rel | 0.73 | 0.13 | 0.40 | 0.63 | 0.71 | 0.84 | 0.95 |
| Avg_Rel | 0.81 | 0.09 | 0.61 | 0.74 | 0.83 | 0.88 | 0.95 |
| Max_Rel | 0.88 | 0.08 | 0.67 | 0.83 | 0.90 | 0.94 | 0.98 |
print(xtable(sum_dat[,c(1:3,5,7)]))% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Wed Feb 16 13:30:50 2022
\begin{table}[ht]
\centering
\begin{tabular}{rrrrrr}
\hline
& avg & sd & min & median & max \\
\hline
SS & 4217.78 & 23241.20 & 36.00 & 603.00 & 263683.00 \\
NLV & 5.46 & 6.93 & 1.00 & 4.00 & 60.00 \\
NOV & 50.12 & 81.58 & 5.00 & 22.50 & 442.00 \\
Min\_Ind & 6.86 & 6.40 & 1.00 & 5.00 & 40.00 \\
Avg\_Ind & 8.95 & 7.00 & 2.50 & 6.22 & 40.00 \\
Max\_Ind & 12.93 & 15.20 & 3.00 & 8.00 & 135.00 \\
Min\_FL & 0.41 & 0.20 & 0.00 & 0.41 & 0.86 \\
Avg\_FL & 0.64 & 0.13 & 0.23 & 0.65 & 0.93 \\
Max\_FL & 0.85 & 0.10 & 0.49 & 0.87 & 1.08 \\
Min\_Rel & 0.73 & 0.13 & 0.40 & 0.71 & 0.95 \\
Avg\_Rel & 0.81 & 0.09 & 0.61 & 0.83 & 0.95 \\
Max\_Rel & 0.88 & 0.08 & 0.67 & 0.90 & 0.98 \\
\hline
\end{tabular}
\end{table}mydata %>%
select(`Scale of Indicators (list)`) %>%
table().
11pt
1
3pt
5
3pt, 4pt
1
3pt, 6pt
1
4pt
22
4pt, 3pt
1
4pt, 5pt
3
4pt, 5pt, 5pt, 5pt, 4pt, 5pt
1
4pt, 6pt, 4pt, 4pt, 4pt, count, sum-scores
1
5pt
42
5pt, 4pt
3
5pt, 4pt, 4pt
1
5pt, 4pt, sum-scores
1
5pt, 5pt
1
5pt, 7pt
6
5pt, 7pt, 4pt
1
5pt, parcels
1
6pt
3
6pt, 5pt, 4pt
1
6pt, 7pt
1
7pt
10
7pt, 5pt, 4pt, dichotomous
2
7pt, dichotomous
1
counts
2
dichotomous
13
dichotomous, 3pt
1
dichotomous, 4pt, 5pt
1
dichotomous, 5pt
1
dichotomous, 5pt, 7pt
1
sum-scores
10
sum-scores, 4pt, 5pt, 7pt
1
sum-scores, dichotomous
1
sum-scores, dichotomous, 5pt, 7pt, force-choice
1 Sample Size
p <- mydata %>%
mutate(`Sample Size` = ifelse(`Sample Size`>5000, 5000, `Sample Size`))%>%
ggplot(aes(x=`Sample Size`))+
geom_histogram(aes(y = ..density..), binwidth = 200)+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 500, color="red")+
geom_vline(xintercept = 2500, color="red")+
scale_x_continuous(breaks=seq(0,5000,500),limits = c(0,5000))+
labs(y=NULL,x=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)
pWarning: Removed 1 rows containing non-finite values (stat_bin).Warning: Removed 1 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).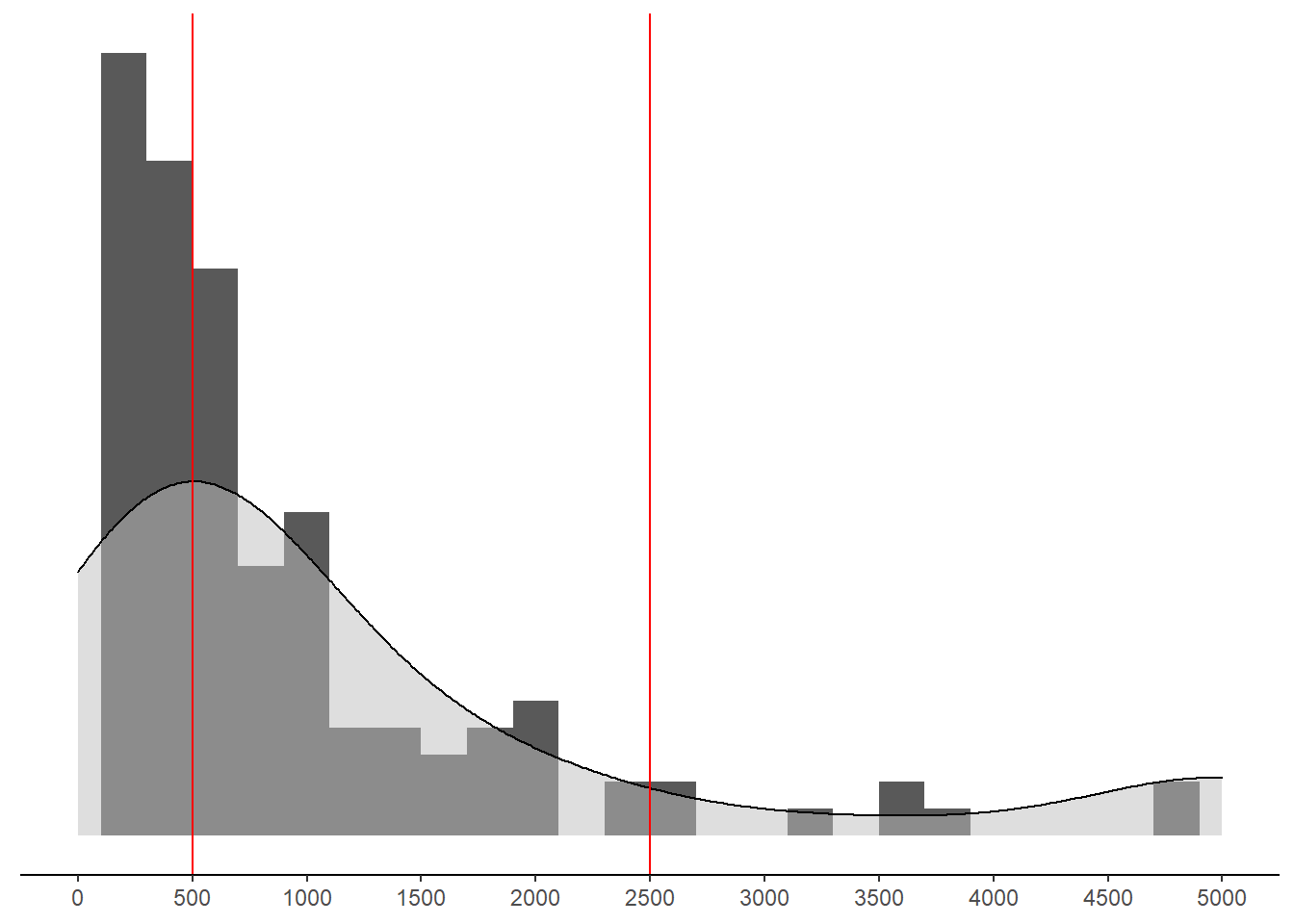
ggsave(filename = "fig/study0_sample_size.pdf",plot=p,width = 4, height=3,units="in")Warning: Removed 1 rows containing non-finite values (stat_bin).Warning: Removed 1 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_sample_size.png",plot=p,width = 4, height=3,units="in")Warning: Removed 1 rows containing non-finite values (stat_bin).Warning: Removed 1 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_sample_size.eps",plot=p,width = 4, height=3,units="in")Warning: Removed 1 rows containing non-finite values (stat_bin).Warning: Removed 1 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).Warning in grid.Call.graphics(C_polygon, x$x, x$y, index): semi-transparency is
not supported on this device: reported only once per pageIndicators
p<-mydata %>%
ggplot(aes(x=`Average Number of Indicators per Factor`))+
geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 5, color="red")+
geom_vline(xintercept = 10, color="red")+
geom_vline(xintercept = 20, color="red")+
scale_x_continuous(breaks=seq(0,40, 5),limits = c(0,45))+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)
pWarning: Removed 4 rows containing non-finite values (stat_bin).Warning: Removed 4 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).
ggsave(filename = "fig/study0_indicators.pdf",plot=p,width = 4, height=3,units="in")Warning: Removed 4 rows containing non-finite values (stat_bin).Warning: Removed 4 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_indicators.png",plot=p,width = 4, height=3,units="in")Warning: Removed 4 rows containing non-finite values (stat_bin).Warning: Removed 4 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_indicators.eps",plot=p,width = 4, height=3,units="in")Warning: Removed 4 rows containing non-finite values (stat_bin).Warning: Removed 4 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).Warning in grid.Call.graphics(C_polygon, x$x, x$y, index): semi-transparency is
not supported on this device: reported only once per pagemydata %>%
ggplot(aes(x=`Minimum Number of Indicators per Factor`))+
geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 5, color="red")+
geom_vline(xintercept = 10, color="red")+
geom_vline(xintercept = 20, color="red")+
scale_x_continuous(breaks=seq(0,40, 5),limits = c(0,45))+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 5 rows containing non-finite values (stat_bin).Warning: Removed 5 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).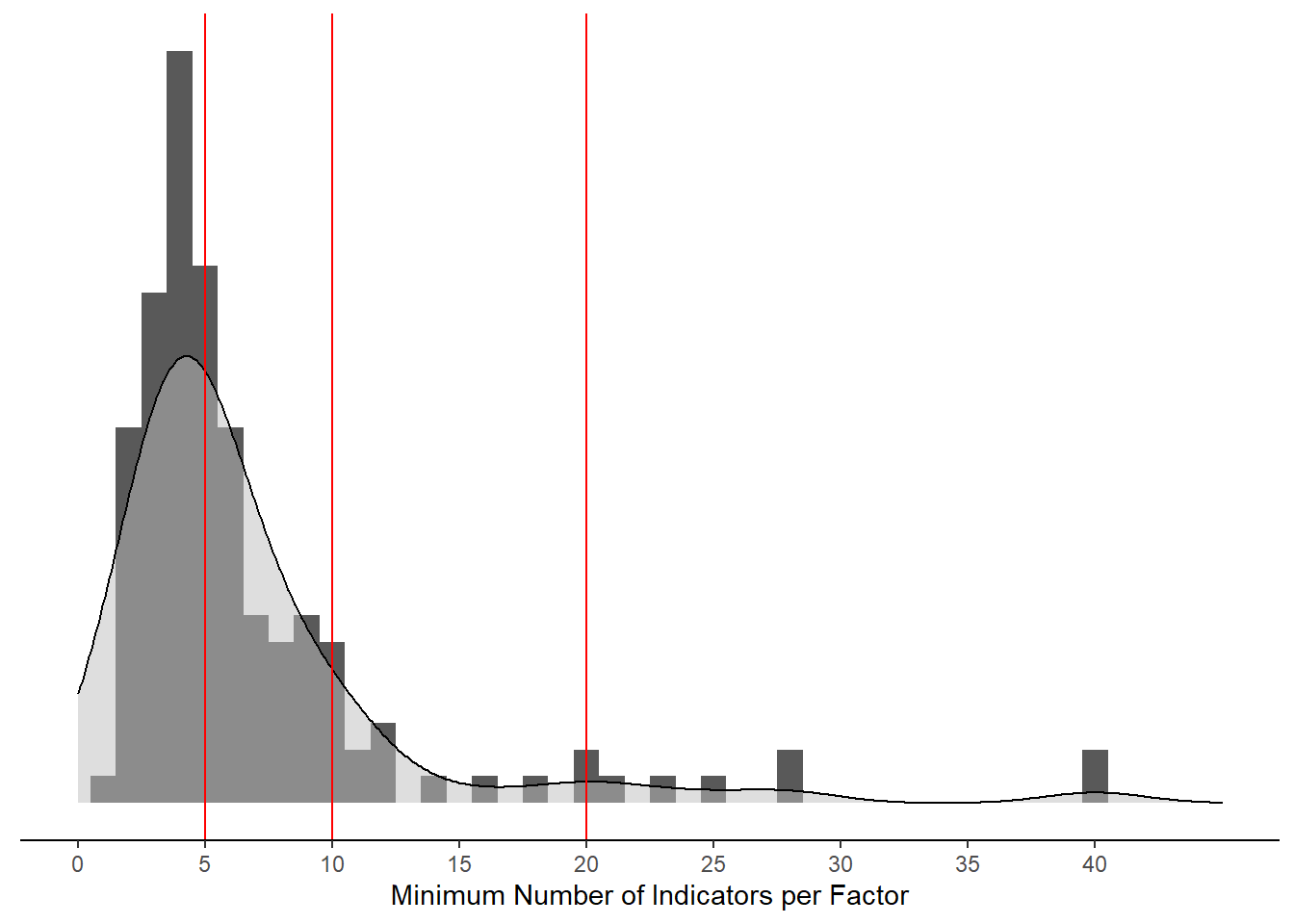
mydata %>%
ggplot(aes(x=`Maximum Number of Indicators per Factor`))+
geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 5, color="red")+
geom_vline(xintercept = 10, color="red")+
geom_vline(xintercept = 20, color="red")+
scale_x_continuous(breaks=seq(0,40, 5),limits = c(0,45))+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 8 rows containing non-finite values (stat_bin).Warning: Removed 8 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).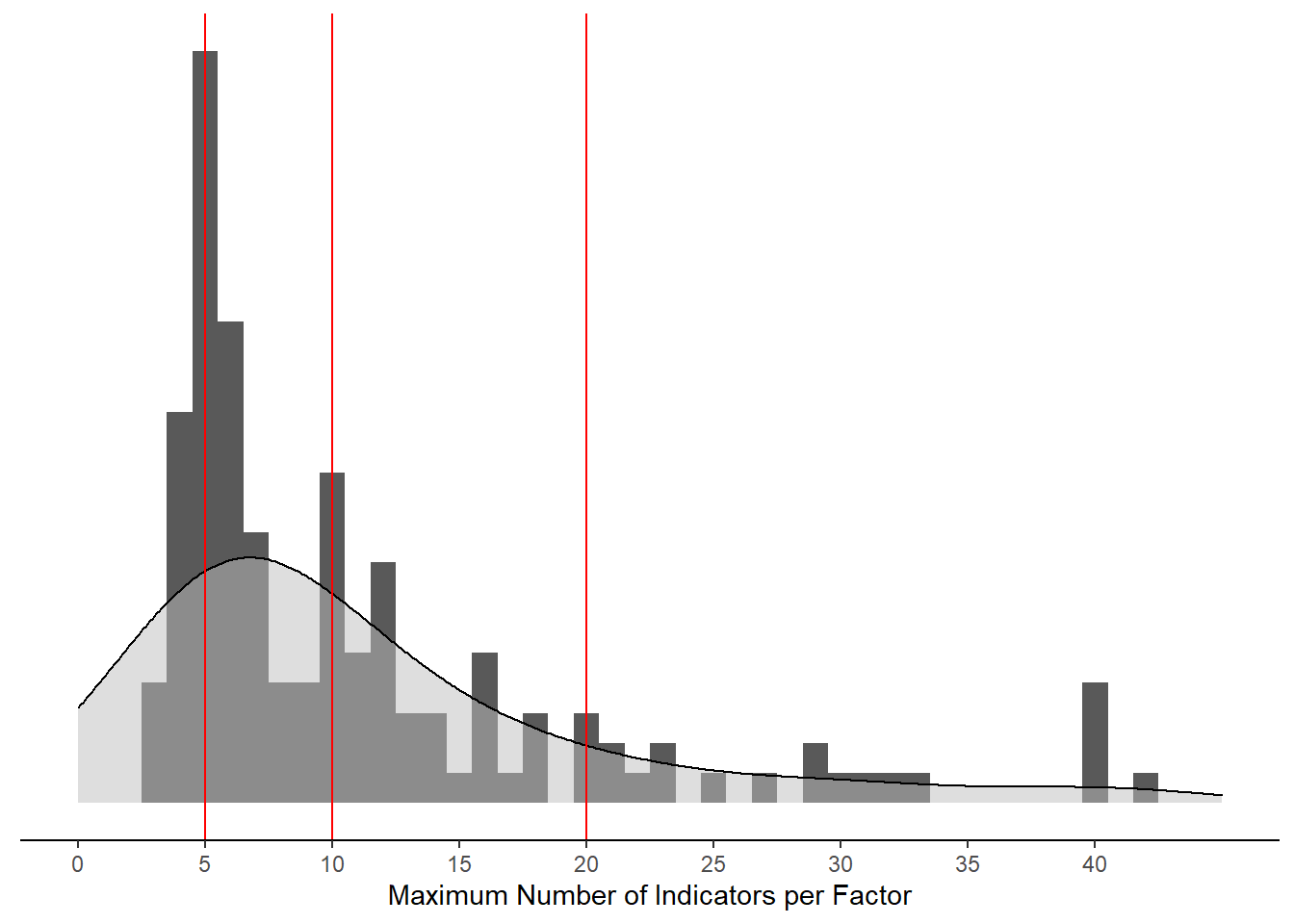
Factor Loadings
mydata %>%
ggplot(aes(x=`Minimum Factor Loading`))+
#geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.9, color="red")+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 49 rows containing non-finite values (stat_density).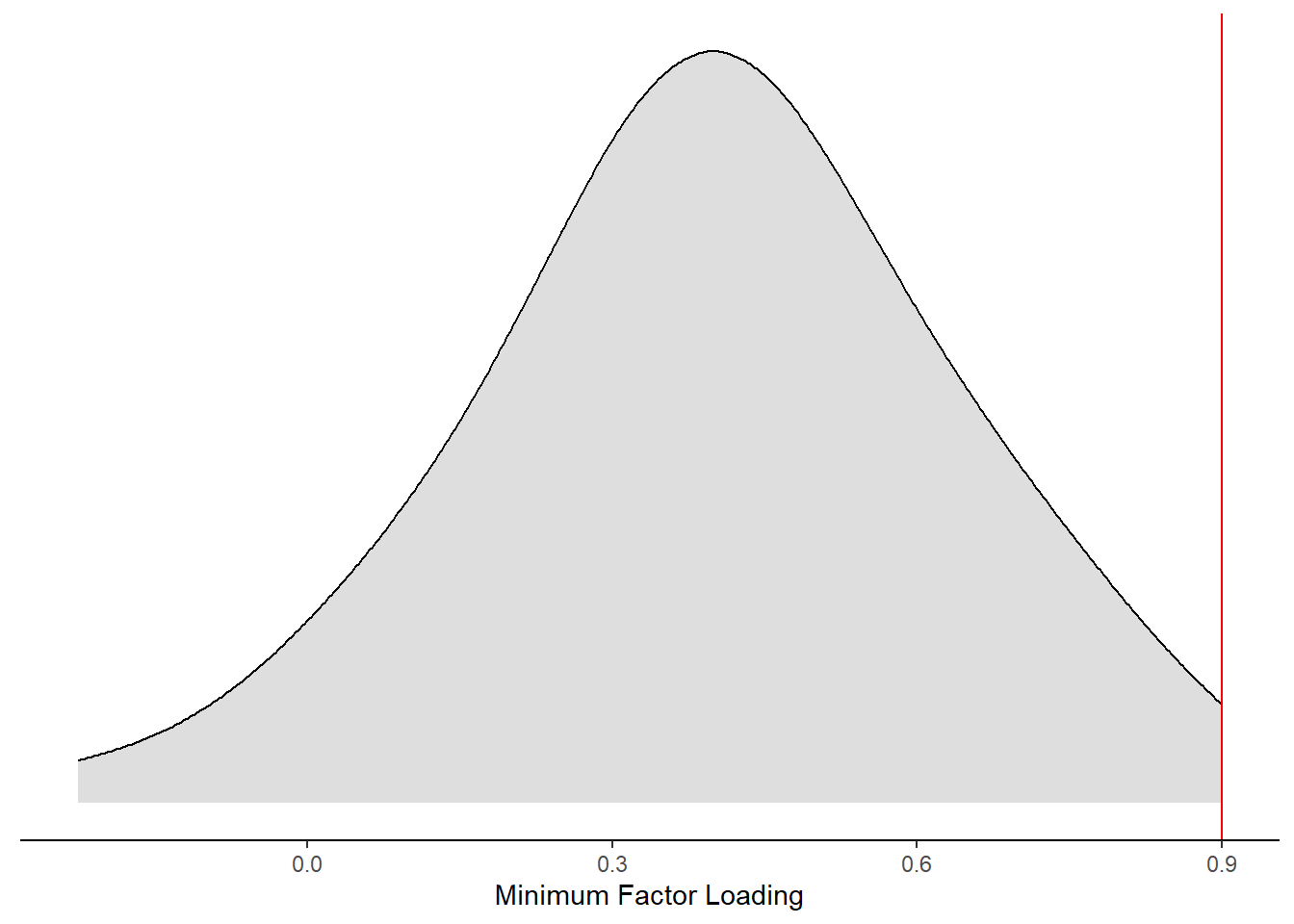
mydata %>%
ggplot(aes(x=`Average Factor Loading`))+
#geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.9, color="red")+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 49 rows containing non-finite values (stat_density).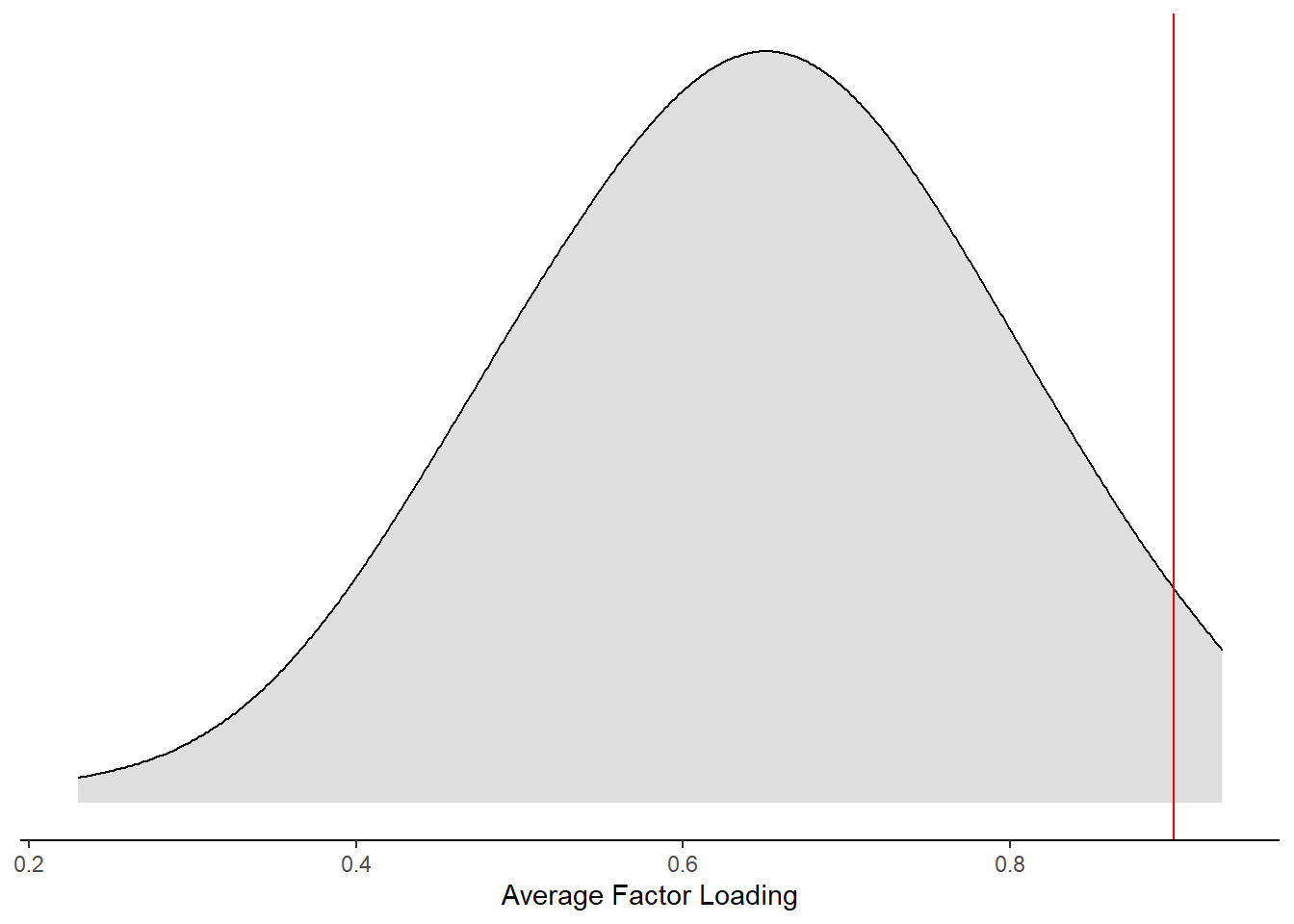
mydata %>%
ggplot(aes(x=`Maximum Factor Loading`))+
#geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.9, color="red")+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 49 rows containing non-finite values (stat_density).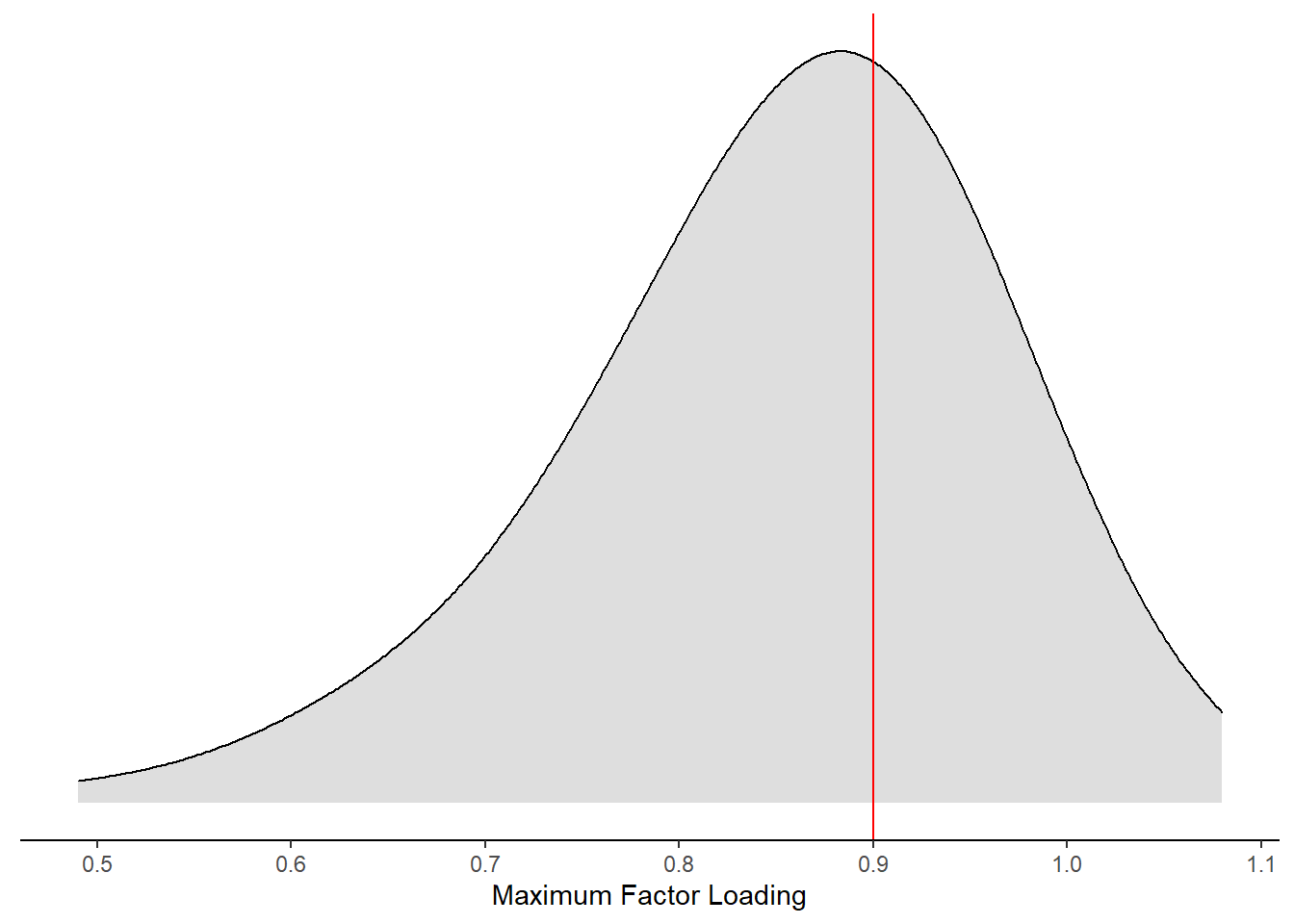
# combine for plot
p<-mydata %>%
select(`Minimum Factor Loading`, `Average Factor Loading`, `Maximum Factor Loading`)%>%
pivot_longer(
cols=everything(),
names_to="type",
values_to="value"
)%>%
ggplot(aes(x=value))+
geom_histogram(binwidth=0.1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.9, color="red")+
scale_x_continuous(breaks=seq(0,1, .25),limits = c(-0.1,1.1))+
labs(y=NULL,x=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)
pWarning: Removed 149 rows containing non-finite values (stat_bin).Warning: Removed 149 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).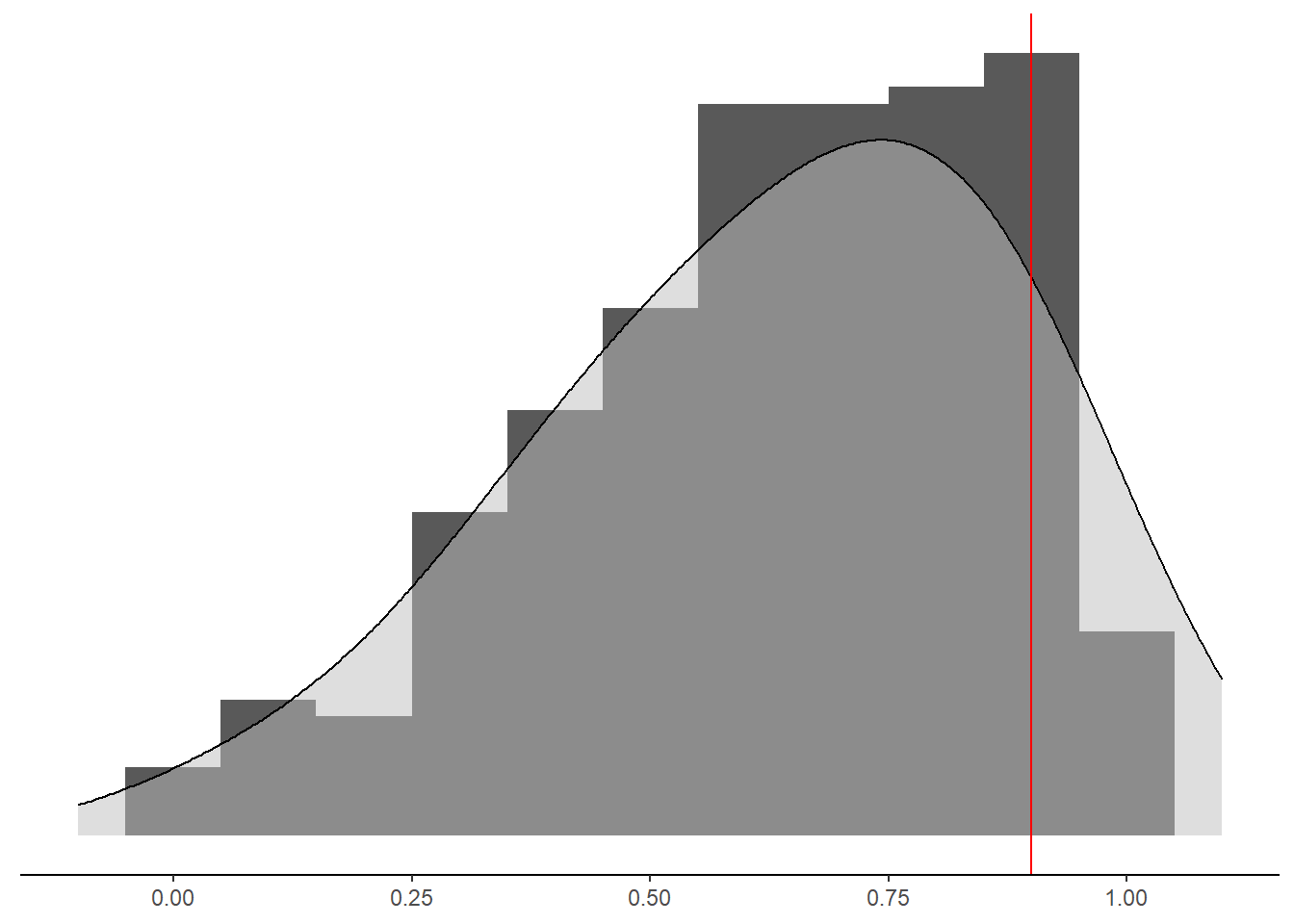
ggsave(filename = "fig/study0_factor_loadings.pdf",plot=p,width = 4, height=3,units="in")Warning: Removed 149 rows containing non-finite values (stat_bin).Warning: Removed 149 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_factor_loadings.png",plot=p,width = 4, height=3,units="in")Warning: Removed 149 rows containing non-finite values (stat_bin).Warning: Removed 149 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_factor_loadings.eps",plot=p,width = 4, height=3,units="in")Warning: Removed 149 rows containing non-finite values (stat_bin).Warning: Removed 149 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).Warning in grid.Call.graphics(C_polygon, x$x, x$y, index): semi-transparency is
not supported on this device: reported only once per pageReliability
mydata %>%
ggplot(aes(x=`Min Reliability`))+
#geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.80, color="red")+
geom_vline(xintercept = 0.89, color="red")+
geom_vline(xintercept = 0.94, color="red")+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 23 rows containing non-finite values (stat_density).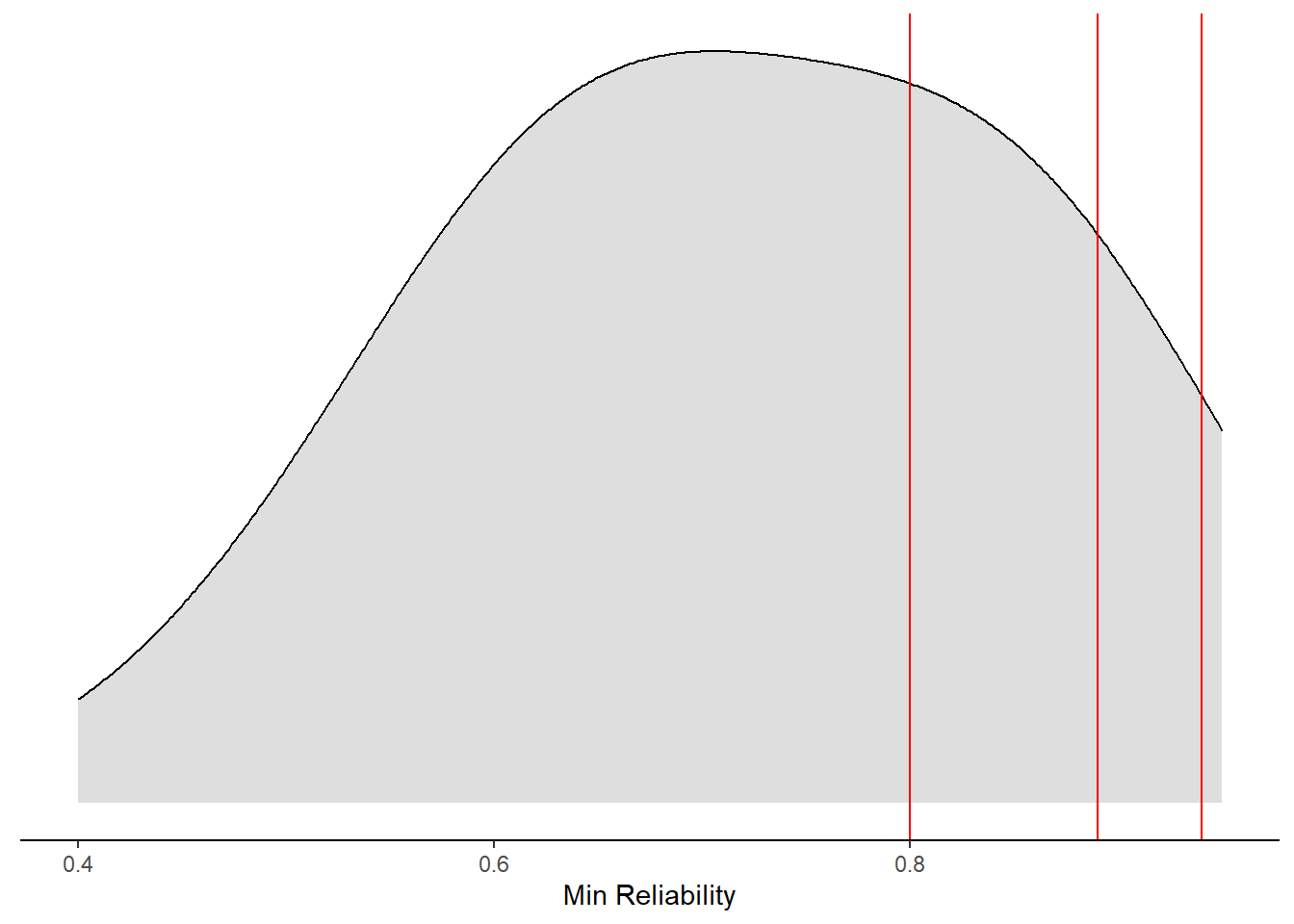
mydata %>%
ggplot(aes(x=`Avg Reliability`))+
#geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.80, color="red")+
geom_vline(xintercept = 0.89, color="red")+
geom_vline(xintercept = 0.94, color="red")+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 23 rows containing non-finite values (stat_density).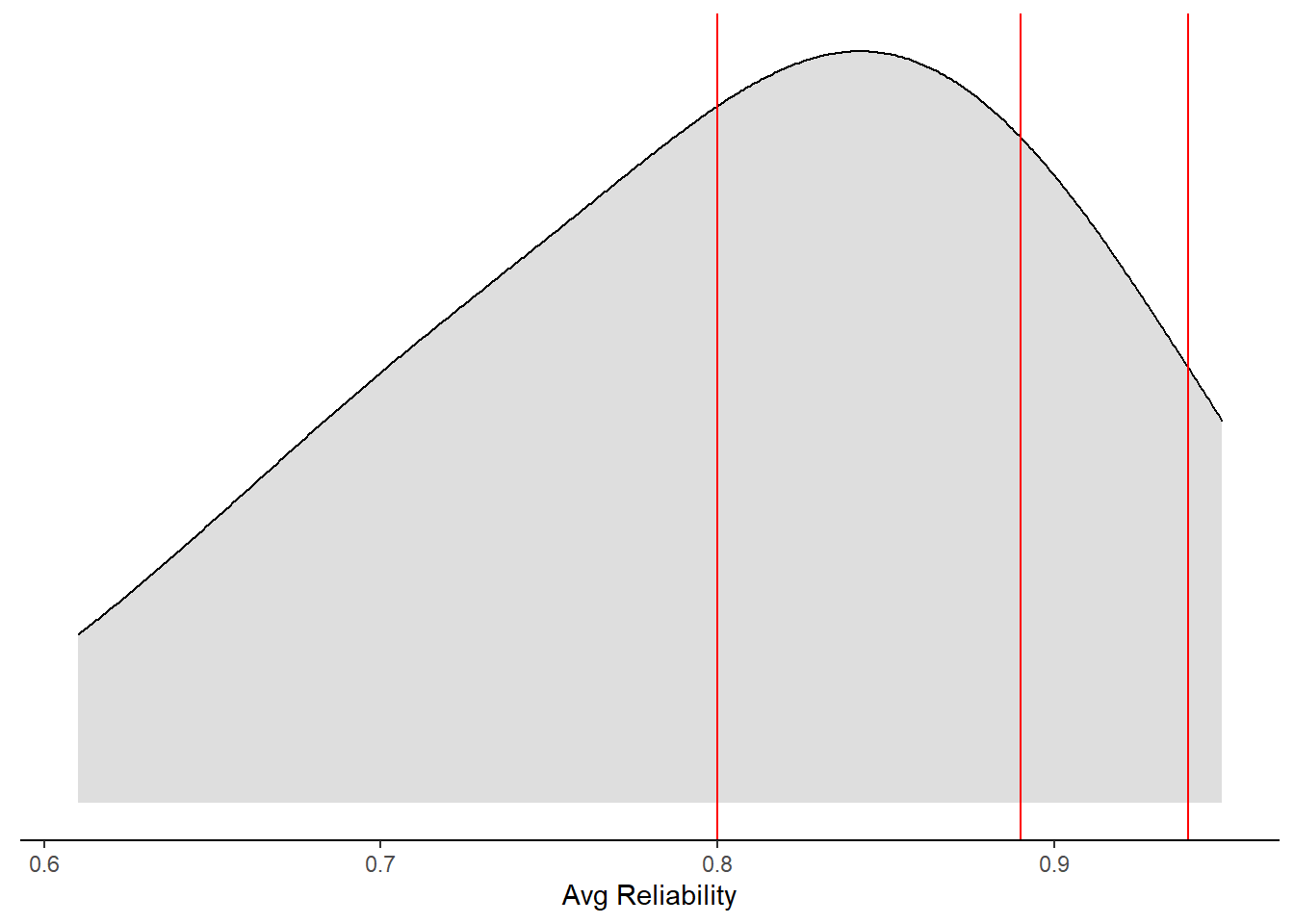
mydata %>%
ggplot(aes(x=`Max Reliability`))+
#geom_histogram(binwidth=1,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.80, color="red")+
geom_vline(xintercept = 0.89, color="red")+
geom_vline(xintercept = 0.94, color="red")+
labs(y=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)Warning: Removed 23 rows containing non-finite values (stat_density).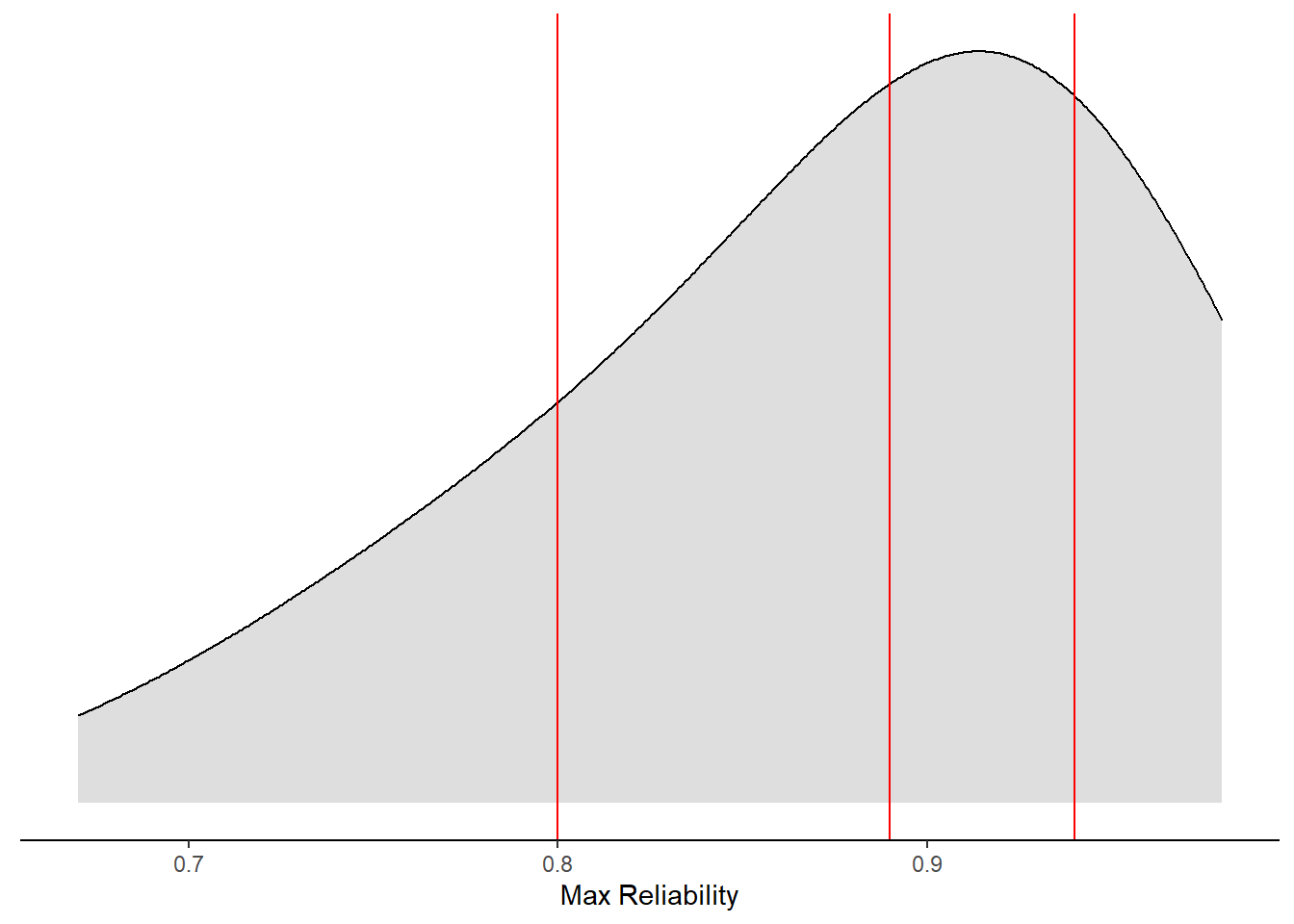
# combine for plot
p<-mydata %>%
select(`Min Reliability`, `Avg Reliability`, `Max Reliability`)%>%
pivot_longer(
cols=everything(),
names_to="type",
values_to="value"
)%>%
ggplot(aes(x=value))+
geom_histogram(binwidth=0.05,aes(y = ..density..))+
geom_density(adjust=2, fill="grey", alpha=0.5)+
geom_vline(xintercept = 0.80, color="red")+
geom_vline(xintercept = 0.89, color="red")+
geom_vline(xintercept = 0.94, color="red")+
scale_x_continuous(breaks=seq(0,1, .25),limits = c(0,1))+
labs(y=NULL,x=NULL)+
theme_classic()+
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y =element_blank()
)
pWarning: Removed 69 rows containing non-finite values (stat_bin).Warning: Removed 69 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).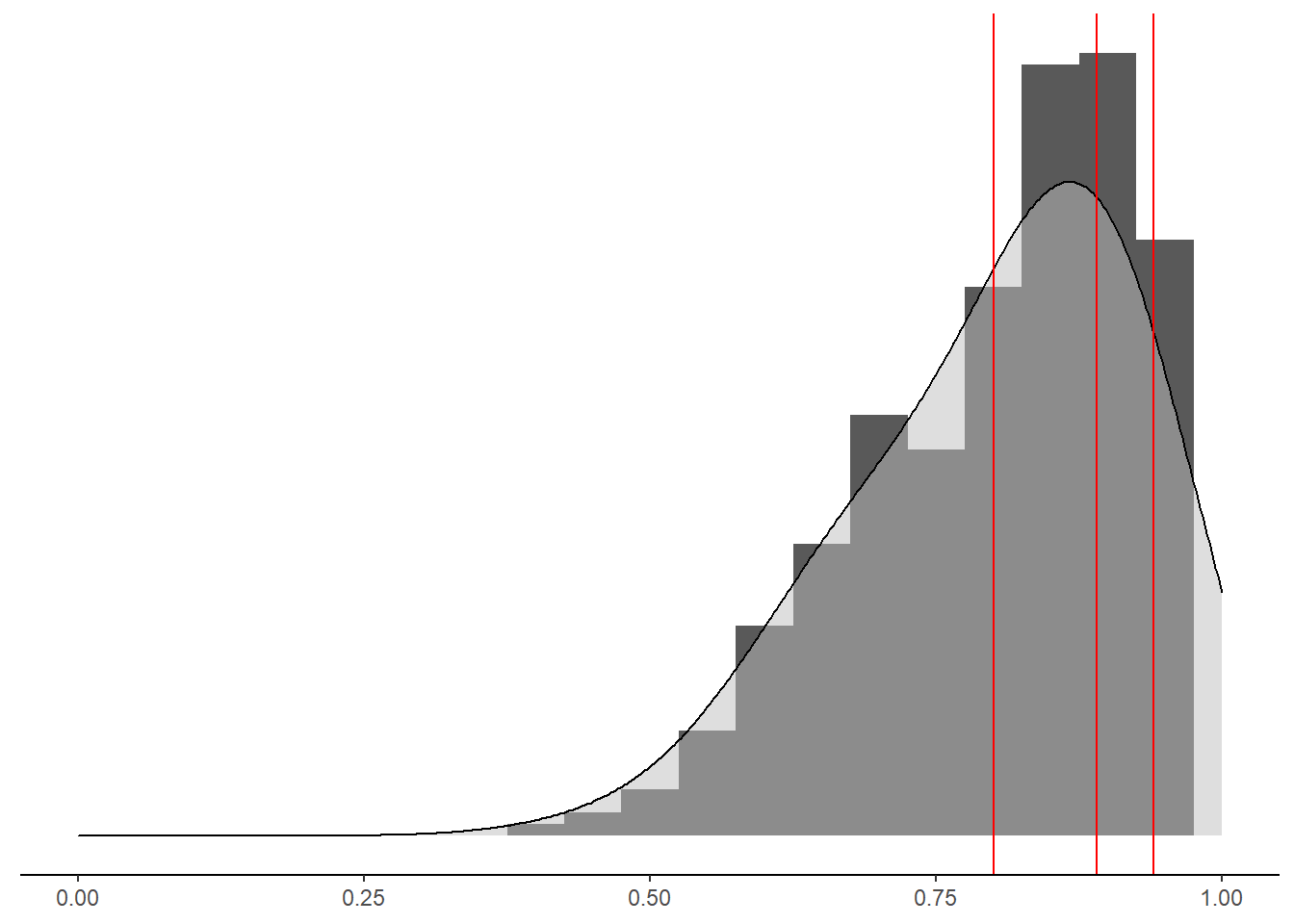
ggsave(filename = "fig/study0_reliability.pdf",plot=p,width = 4, height=3,units="in")Warning: Removed 69 rows containing non-finite values (stat_bin).Warning: Removed 69 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_reliability.png",plot=p,width = 4, height=3,units="in")Warning: Removed 69 rows containing non-finite values (stat_bin).Warning: Removed 69 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).ggsave(filename = "fig/study0_reliability.eps",plot=p,width = 4, height=3,units="in")Warning: Removed 69 rows containing non-finite values (stat_bin).Warning: Removed 69 rows containing non-finite values (stat_density).Warning: Removed 2 rows containing missing values (geom_bar).Warning in grid.Call.graphics(C_polygon, x$x, x$y, index): semi-transparency is
not supported on this device: reported only once per pagesum(mydata$`Method of Computing Reliability` == "omega", na.rm=T)/nrow(mydata)[1] 0.16Response Categories
mydata %>%
select(`Scale of Indicators (list)`) %>%
table().
11pt
1
3pt
5
3pt, 4pt
1
3pt, 6pt
1
4pt
22
4pt, 3pt
1
4pt, 5pt
3
4pt, 5pt, 5pt, 5pt, 4pt, 5pt
1
4pt, 6pt, 4pt, 4pt, 4pt, count, sum-scores
1
5pt
42
5pt, 4pt
3
5pt, 4pt, 4pt
1
5pt, 4pt, sum-scores
1
5pt, 5pt
1
5pt, 7pt
6
5pt, 7pt, 4pt
1
5pt, parcels
1
6pt
3
6pt, 5pt, 4pt
1
6pt, 7pt
1
7pt
10
7pt, 5pt, 4pt, dichotomous
2
7pt, dichotomous
1
counts
2
dichotomous
13
dichotomous, 3pt
1
dichotomous, 4pt, 5pt
1
dichotomous, 5pt
1
dichotomous, 5pt, 7pt
1
sum-scores
10
sum-scores, 4pt, 5pt, 7pt
1
sum-scores, dichotomous
1
sum-scores, dichotomous, 5pt, 7pt, force-choice
1 indCats <- mydata$`Scale of Indicators (list)`
ind2 <- ind3 <- ind4 <- ind5 <- ind6 <- ind7 <- indSS <- numeric(length(indCats))
for(i in 1:length(indCats)){
indi <- str_split(indCats[i], ",", simplify = T)
ind2[i] <- ifelse(any(indi %like% "dichotomous"), 1, 0)
ind3[i] <- ifelse(any(indi %like% "3pt"), 1, 0)
ind4[i] <- ifelse(any(indi %like% "4pt"), 1, 0)
ind5[i] <- ifelse(any(indi %like% "5pt"), 1, 0)
ind6[i] <- ifelse(any(indi %like% "6pt"), 1, 0)
ind7[i] <- ifelse(any(indi %like% "7pt"), 1, 0)
indSS[i] <- ifelse(any(indi %like% "sum-score"), 1, 0)
}
catDat <- as.data.frame(cbind(ind2, ind3, ind4, ind5, ind6, ind7, indSS))
catDat$multi <- ifelse(rowSums(catDat) > 1, 1, 0)
# summarizing
mean(ind2)[1] 0.153mean(ind3)[1] 0.0625mean(ind4)[1] 0.278mean(ind5)[1] 0.472mean(ind6)[1] 0.0486mean(ind7)[1] 0.167mean(indSS)[1] 0.104mean(catDat$multi)[1] 0.222
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 car_3.0-10 carData_3.0-4
[4] mvtnorm_1.1-1 LaplacesDemon_16.1.4 runjags_2.2.0-2
[7] lme4_1.1-26 Matrix_1.3-2 sirt_3.9-4
[10] R2jags_0.6-1 rjags_4-12 eRm_1.0-2
[13] diffIRT_1.5 statmod_1.4.35 xtable_1.8-4
[16] kableExtra_1.3.4 lavaan_0.6-7 polycor_0.7-10
[19] bayesplot_1.8.0 ggmcmc_1.5.1.1 coda_0.19-4
[22] data.table_1.14.0 patchwork_1.1.1 forcats_0.5.1
[25] stringr_1.4.0 dplyr_1.0.5 purrr_0.3.4
[28] readr_1.4.0 tidyr_1.1.3 tibble_3.1.0
[31] ggplot2_3.3.5 tidyverse_1.3.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] minqa_1.2.4 TAM_3.5-19 colorspace_2.0-0 rio_0.5.26
[5] ellipsis_0.3.1 ggridges_0.5.3 rprojroot_2.0.2 fs_1.5.0
[9] rstudioapi_0.13 farver_2.1.0 fansi_0.4.2 lubridate_1.7.10
[13] xml2_1.3.2 splines_4.0.5 mnormt_2.0.2 knitr_1.31
[17] jsonlite_1.7.2 nloptr_1.2.2.2 broom_0.7.5 dbplyr_2.1.0
[21] compiler_4.0.5 httr_1.4.2 backports_1.2.1 assertthat_0.2.1
[25] cli_2.3.1 later_1.1.0.1 htmltools_0.5.1.1 tools_4.0.5
[29] gtable_0.3.0 glue_1.4.2 Rcpp_1.0.7 cellranger_1.1.0
[33] jquerylib_0.1.3 vctrs_0.3.6 svglite_2.0.0 nlme_3.1-152
[37] psych_2.0.12 xfun_0.21 ps_1.6.0 openxlsx_4.2.3
[41] rvest_1.0.0 lifecycle_1.0.0 MASS_7.3-53.1 scales_1.1.1
[45] ragg_1.1.1 hms_1.0.0 promises_1.2.0.1 parallel_4.0.5
[49] RColorBrewer_1.1-2 curl_4.3 yaml_2.2.1 sass_0.3.1
[53] reshape_0.8.8 stringi_1.5.3 highr_0.8 zip_2.1.1
[57] boot_1.3-27 rlang_0.4.10 pkgconfig_2.0.3 systemfonts_1.0.1
[61] evaluate_0.14 lattice_0.20-41 labeling_0.4.2 tidyselect_1.1.0
[65] GGally_2.1.1 plyr_1.8.6 magrittr_2.0.1 R6_2.5.0
[69] generics_0.1.0 DBI_1.1.1 foreign_0.8-81 pillar_1.5.1
[73] haven_2.3.1 withr_2.4.1 abind_1.4-5 modelr_0.1.8
[77] crayon_1.4.1 utf8_1.1.4 tmvnsim_1.0-2 rmarkdown_2.7
[81] grid_4.0.5 CDM_7.5-15 pbivnorm_0.6.0 git2r_0.28.0
[85] reprex_1.0.0 digest_0.6.27 webshot_0.5.2 httpuv_1.5.5
[89] textshaping_0.3.1 stats4_4.0.5 munsell_0.5.0 viridisLite_0.3.0
[93] bslib_0.2.4 R2WinBUGS_2.1-21