Simulation Study 2
Posterior Convergence
R. Noah Padgett
2022-01-09
Last updated: 2022-02-13
Checks: 5 1
Knit directory: Padgett-Dissertation/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210401) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Tracking code development and connecting the code version to the results is critical for reproducibility. To start using Git, open the Terminal and type git init in your project directory.
This project is not being versioned with Git. To obtain the full reproducibility benefits of using workflowr, please see ?wflow_start.
# Load packages
source("code/load_packages.R")
source("code/load_utility_functions.R")
# environment options
options(scipen = 999, digits=3)
# results folder
res_folder <- paste0(w.d, "/data/study_2")
# read in data
source("code/load_extracted_data.R")Convergence
This is component of the project, a coarse grain investigation of the posteriors’ convergence will be assessed. In the proposal, I stated that I will evaluate convergence of the posteriors using the R-hat value as measures of convergence. R-hat values close to 1, within 0.1, will be considered a “converged” posterior for the parameter.
The code below loops through the posterior summaries to identify converged solutions.
mydata <- mydata %>%
# group by condition, iteration, and parameter group to get Rhat more summarised
group_by(condition, iter, parameter_group) %>%
mutate(Rhat_paragrp = mean(Rhat)) %>%
ungroup() %>%
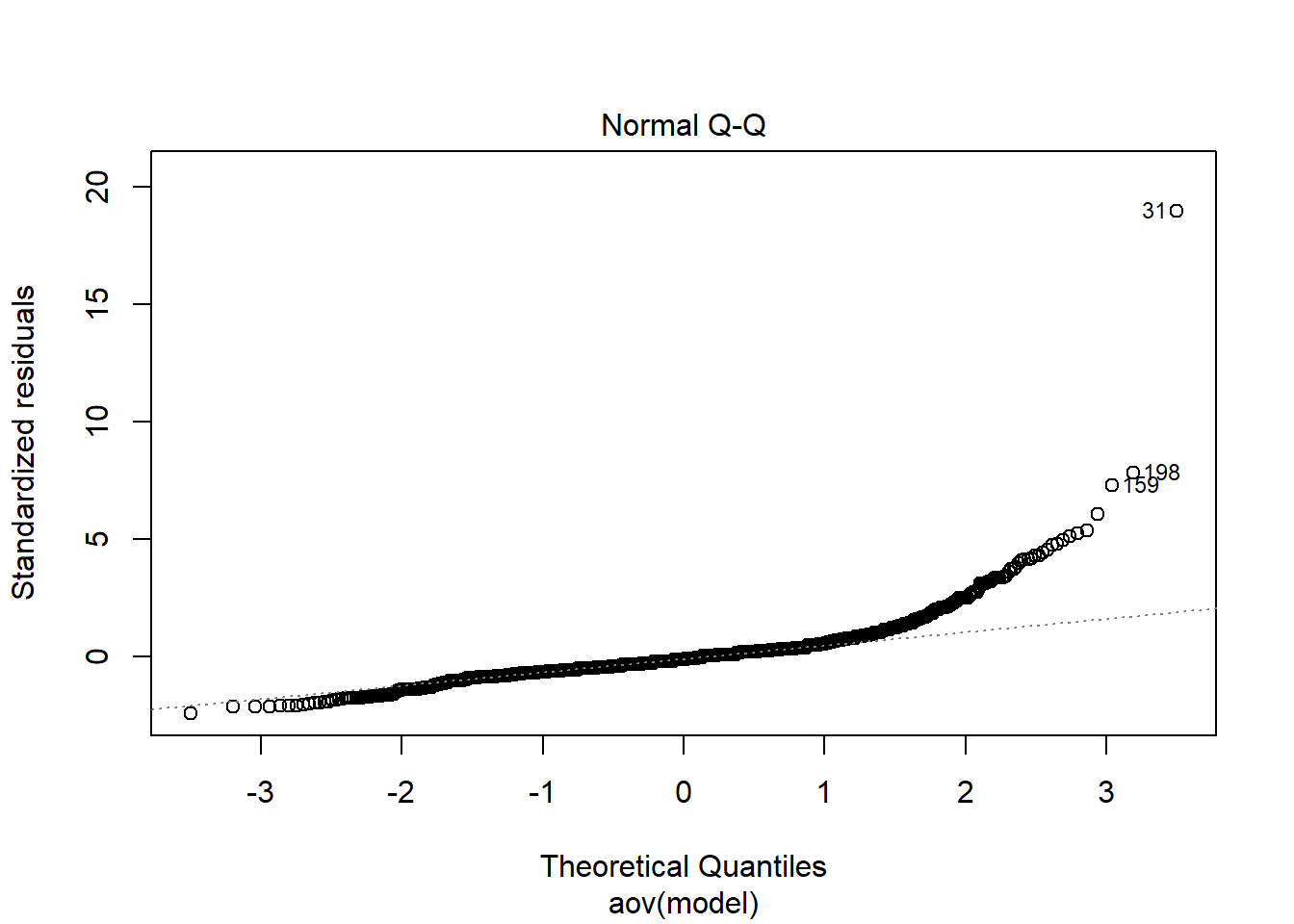
mutate(
converge = ifelse(Rhat - 1.10 < 0, 1, 0),
converge_paragrp = ifelse(Rhat_paragrp - 1.10 < 0, 1, 0)
) %>%
group_by(condition, iter, parameter_group) %>%
mutate(
converge_proportion_paragrp = sum(converge)/n(),
)
# Update condition labels for plotting
mydata$condition_lbs <- factor(
mydata$condition,
levels = 1:24,
labels = c(
"nCat=2, nItem= 5, N= 500",
"nCat=2, nItem= 5, N=2500",
"nCat=2, nItem=10, N= 500",
"nCat=2, nItem=10, N=2500",
"nCat=2, nItem=20, N= 500",
"nCat=2, nItem=20, N=2500",
"nCat=5, nItem= 5, N= 500",
"nCat=5, nItem= 5, N=2500",
"nCat=5, nItem=10, N= 500",
"nCat=5, nItem=10, N=2500",
"nCat=5, nItem=20, N= 500",
"nCat=5, nItem=20, N=2500",
"nCat=3, nItem= 5, N= 500",
"nCat=3, nItem= 5, N=2500",
"nCat=3, nItem=10, N= 500",
"nCat=3, nItem=10, N=2500",
"nCat=3, nItem=20, N= 500",
"nCat=3, nItem=20, N=2500",
"nCat=7, nItem= 5, N= 500",
"nCat=7, nItem= 5, N=2500",
"nCat=7, nItem=10, N= 500",
"nCat=7, nItem=10, N=2500",
"nCat=7, nItem=20, N= 500",
"nCat=7, nItem=20, N=2500"
)
)
# columns to keep
keep_cols <- c("condition","condition_lbs", "iter", "N", "N_items", "N_cat", "parameter_group", "Rhat_paragrp", "converge_paragrp")Summaring Convergence
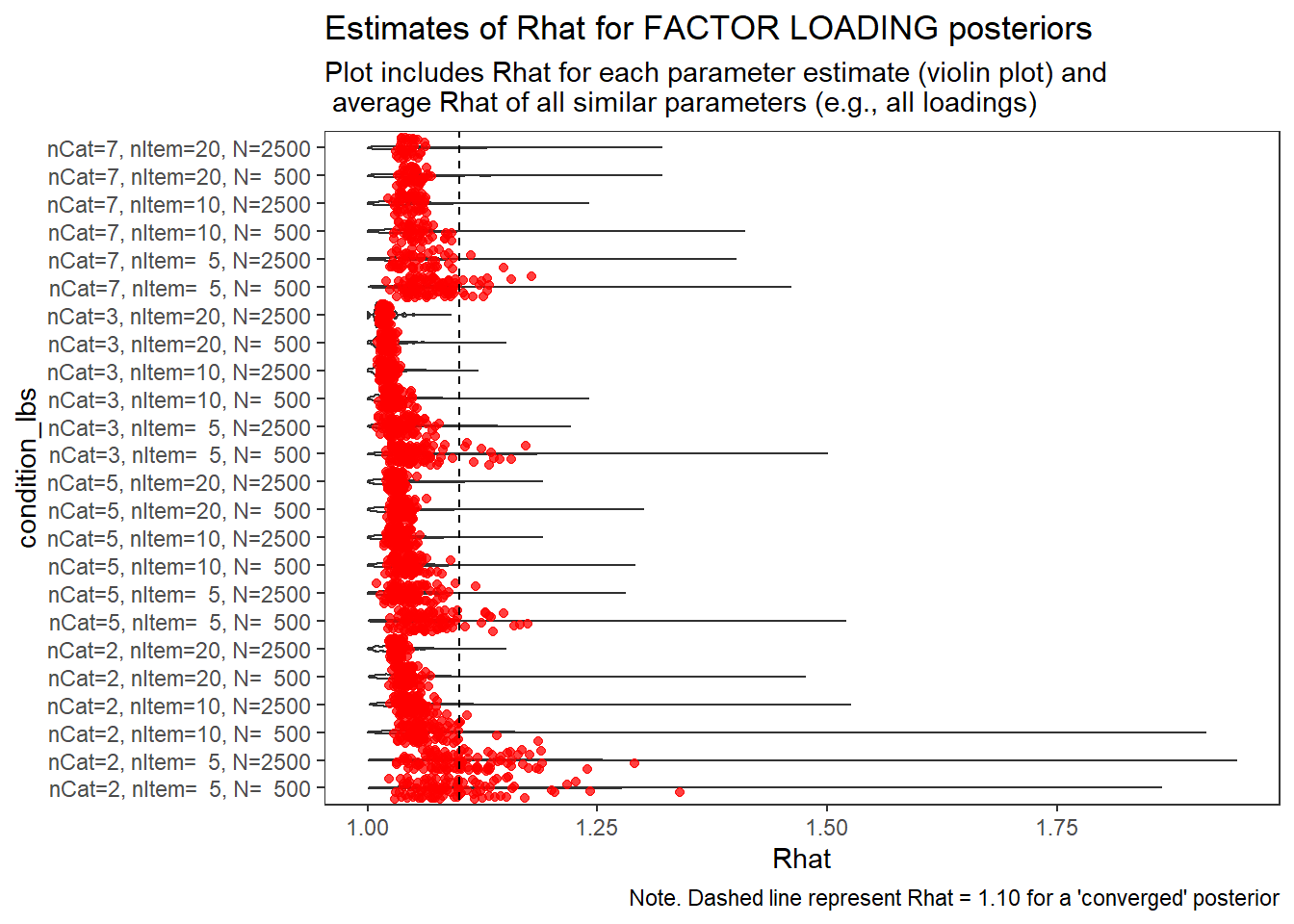
Factor Loadings (\(\lambda\))
param = "FACTOR LOADING"
conv <- mydata %>%
filter(parameter_group == "lambda")
conv_para_grp <- mydata %>%
filter(parameter_group == "lambda") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 500 | 341 | 0.682 |
| 2 | 2 | 5 | 2500 | 500 | 327 | 0.654 |
| 3 | 2 | 10 | 500 | 1000 | 846 | 0.846 |
| 4 | 2 | 10 | 2500 | 1000 | 900 | 0.900 |
| 5 | 2 | 20 | 500 | 2000 | 1898 | 0.949 |
| 6 | 2 | 20 | 2500 | 2000 | 1945 | 0.973 |
| 7 | 5 | 5 | 500 | 500 | 388 | 0.776 |
| 8 | 5 | 5 | 2500 | 500 | 456 | 0.912 |
| 9 | 5 | 10 | 500 | 1000 | 920 | 0.920 |
| 10 | 5 | 10 | 2500 | 1000 | 965 | 0.965 |
| 11 | 5 | 20 | 500 | 2000 | 1890 | 0.945 |
| 12 | 5 | 20 | 2500 | 2000 | 1933 | 0.967 |
| 13 | 3 | 5 | 500 | 500 | 433 | 0.866 |
| 14 | 3 | 5 | 2500 | 500 | 474 | 0.948 |
| 15 | 3 | 10 | 500 | 1000 | 980 | 0.980 |
| 16 | 3 | 10 | 2500 | 1000 | 995 | 0.995 |
| 17 | 3 | 20 | 500 | 2000 | 1987 | 0.994 |
| 18 | 3 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 19 | 7 | 5 | 500 | 500 | 389 | 0.778 |
| 20 | 7 | 5 | 2500 | 250 | 207 | 0.828 |
| 21 | 7 | 10 | 500 | 500 | 424 | 0.848 |
| 22 | 7 | 10 | 2500 | 500 | 451 | 0.902 |
| 23 | 7 | 20 | 500 | 1000 | 879 | 0.879 |
| 24 | 7 | 20 | 2500 | 760 | 700 | 0.921 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 69 | 0.69 |
| 2 | 2 | 5 | 2500 | 100 | 56 | 0.56 |
| 3 | 2 | 10 | 500 | 100 | 97 | 0.97 |
| 4 | 2 | 10 | 2500 | 100 | 99 | 0.99 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 89 | 0.89 |
| 8 | 5 | 5 | 2500 | 100 | 99 | 0.99 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 90 | 0.90 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 87 | 0.87 |
| 20 | 7 | 5 | 2500 | 50 | 48 | 0.96 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
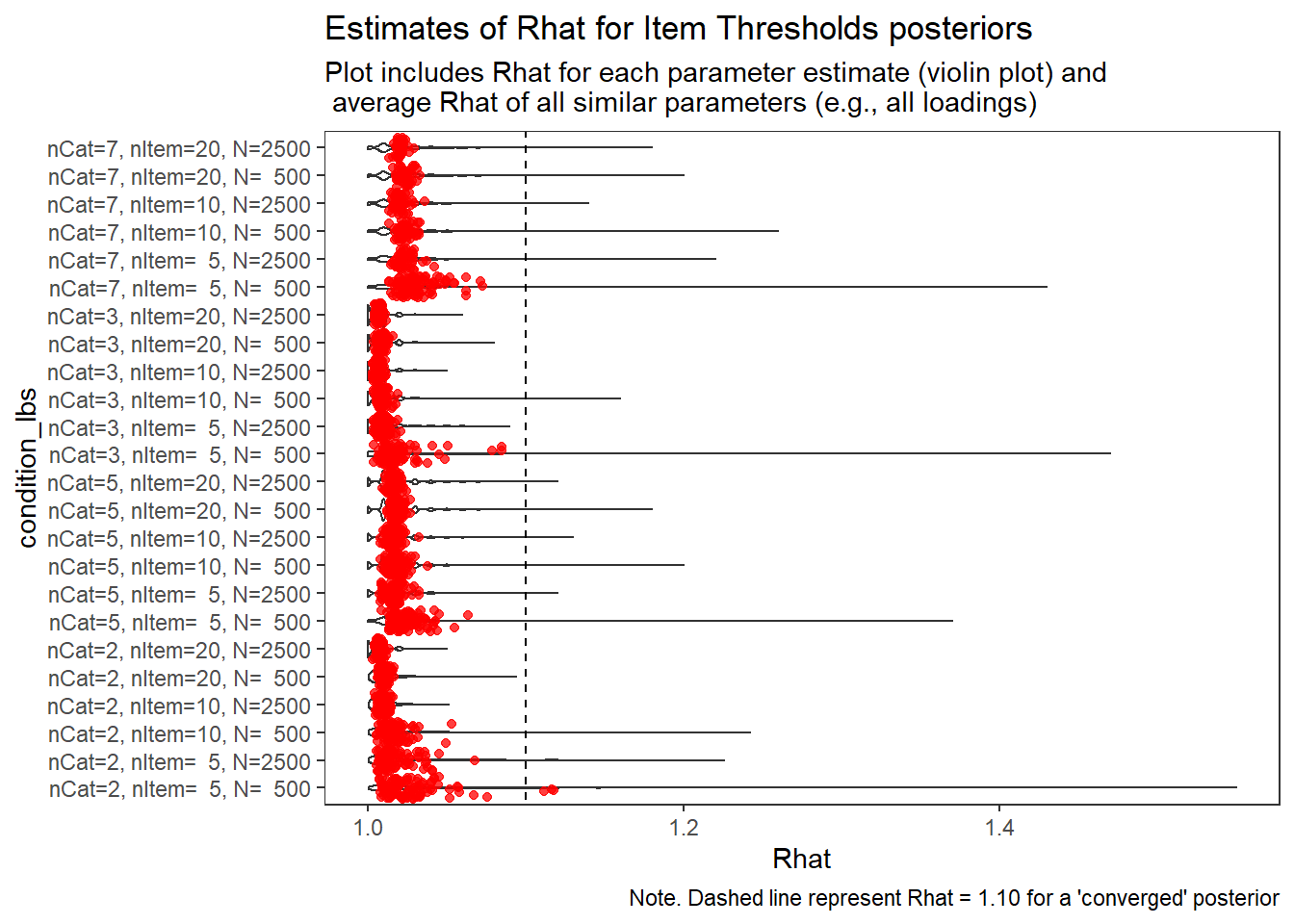
Item Thresholds (\(\tau\))
param = "Item Thresholds"
conv <- mydata %>%
filter(parameter_group == "tau")
conv_para_grp <- mydata %>%
filter(parameter_group == "tau") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 500 | 478 | 0.956 |
| 2 | 2 | 5 | 2500 | 500 | 491 | 0.982 |
| 3 | 2 | 10 | 500 | 1000 | 985 | 0.985 |
| 4 | 2 | 10 | 2500 | 1000 | 1000 | 1.000 |
| 5 | 2 | 20 | 500 | 2000 | 2000 | 1.000 |
| 6 | 2 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 7 | 5 | 5 | 500 | 2000 | 1943 | 0.972 |
| 8 | 5 | 5 | 2500 | 2000 | 1997 | 0.999 |
| 9 | 5 | 10 | 500 | 4000 | 3980 | 0.995 |
| 10 | 5 | 10 | 2500 | 4000 | 3998 | 1.000 |
| 11 | 5 | 20 | 500 | 8000 | 7970 | 0.996 |
| 12 | 5 | 20 | 2500 | 8000 | 7990 | 0.999 |
| 13 | 3 | 5 | 500 | 1000 | 977 | 0.977 |
| 14 | 3 | 5 | 2500 | 1000 | 1000 | 1.000 |
| 15 | 3 | 10 | 500 | 2000 | 1997 | 0.999 |
| 16 | 3 | 10 | 2500 | 2000 | 2000 | 1.000 |
| 17 | 3 | 20 | 500 | 4000 | 4000 | 1.000 |
| 18 | 3 | 20 | 2500 | 4000 | 4000 | 1.000 |
| 19 | 7 | 5 | 500 | 3000 | 2854 | 0.951 |
| 20 | 7 | 5 | 2500 | 1500 | 1483 | 0.989 |
| 21 | 7 | 10 | 500 | 3000 | 2971 | 0.990 |
| 22 | 7 | 10 | 2500 | 3000 | 2989 | 0.996 |
| 23 | 7 | 20 | 500 | 6000 | 5933 | 0.989 |
| 24 | 7 | 20 | 2500 | 4560 | 4538 | 0.995 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 97 | 0.97 |
| 2 | 2 | 5 | 2500 | 100 | 100 | 1.00 |
| 3 | 2 | 10 | 500 | 100 | 100 | 1.00 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1.00 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 100 | 1.00 |
| 8 | 5 | 5 | 2500 | 100 | 100 | 1.00 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 100 | 1.00 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 100 | 1.00 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1.00 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
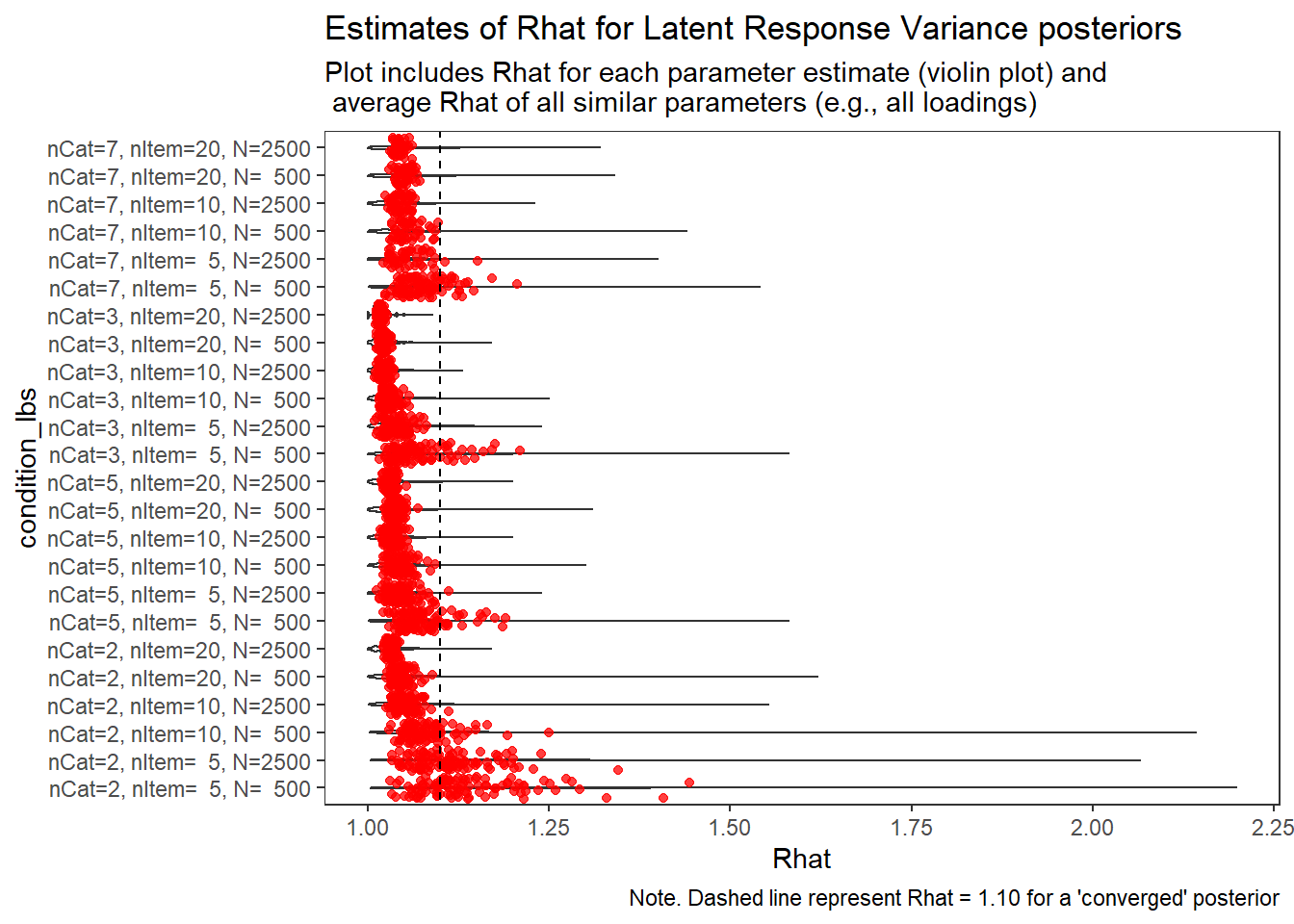
Latent Response Variance (\(\theta\))
param = "Latent Response Variance"
conv <- mydata %>%
filter(parameter_group == "theta")
conv_para_grp <- mydata %>%
filter(parameter_group == "theta") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 500 | 250 | 0.500 |
| 2 | 2 | 5 | 2500 | 500 | 310 | 0.620 |
| 3 | 2 | 10 | 500 | 1000 | 756 | 0.756 |
| 4 | 2 | 10 | 2500 | 1000 | 887 | 0.887 |
| 5 | 2 | 20 | 500 | 2000 | 1844 | 0.922 |
| 6 | 2 | 20 | 2500 | 2000 | 1941 | 0.971 |
| 7 | 5 | 5 | 500 | 500 | 366 | 0.732 |
| 8 | 5 | 5 | 2500 | 500 | 452 | 0.904 |
| 9 | 5 | 10 | 500 | 1000 | 908 | 0.908 |
| 10 | 5 | 10 | 2500 | 1000 | 964 | 0.964 |
| 11 | 5 | 20 | 500 | 2000 | 1876 | 0.938 |
| 12 | 5 | 20 | 2500 | 2000 | 1932 | 0.966 |
| 13 | 3 | 5 | 500 | 500 | 402 | 0.804 |
| 14 | 3 | 5 | 2500 | 500 | 465 | 0.930 |
| 15 | 3 | 10 | 500 | 1000 | 976 | 0.976 |
| 16 | 3 | 10 | 2500 | 1000 | 993 | 0.993 |
| 17 | 3 | 20 | 500 | 2000 | 1986 | 0.993 |
| 18 | 3 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 19 | 7 | 5 | 500 | 500 | 374 | 0.748 |
| 20 | 7 | 5 | 2500 | 250 | 205 | 0.820 |
| 21 | 7 | 10 | 500 | 500 | 420 | 0.840 |
| 22 | 7 | 10 | 2500 | 500 | 451 | 0.902 |
| 23 | 7 | 20 | 500 | 1000 | 861 | 0.861 |
| 24 | 7 | 20 | 2500 | 760 | 700 | 0.921 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 33 | 0.33 |
| 2 | 2 | 5 | 2500 | 100 | 46 | 0.46 |
| 3 | 2 | 10 | 500 | 100 | 85 | 0.85 |
| 4 | 2 | 10 | 2500 | 100 | 99 | 0.99 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 82 | 0.82 |
| 8 | 5 | 5 | 2500 | 100 | 99 | 0.99 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 82 | 0.82 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 79 | 0.79 |
| 20 | 7 | 5 | 2500 | 50 | 48 | 0.96 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
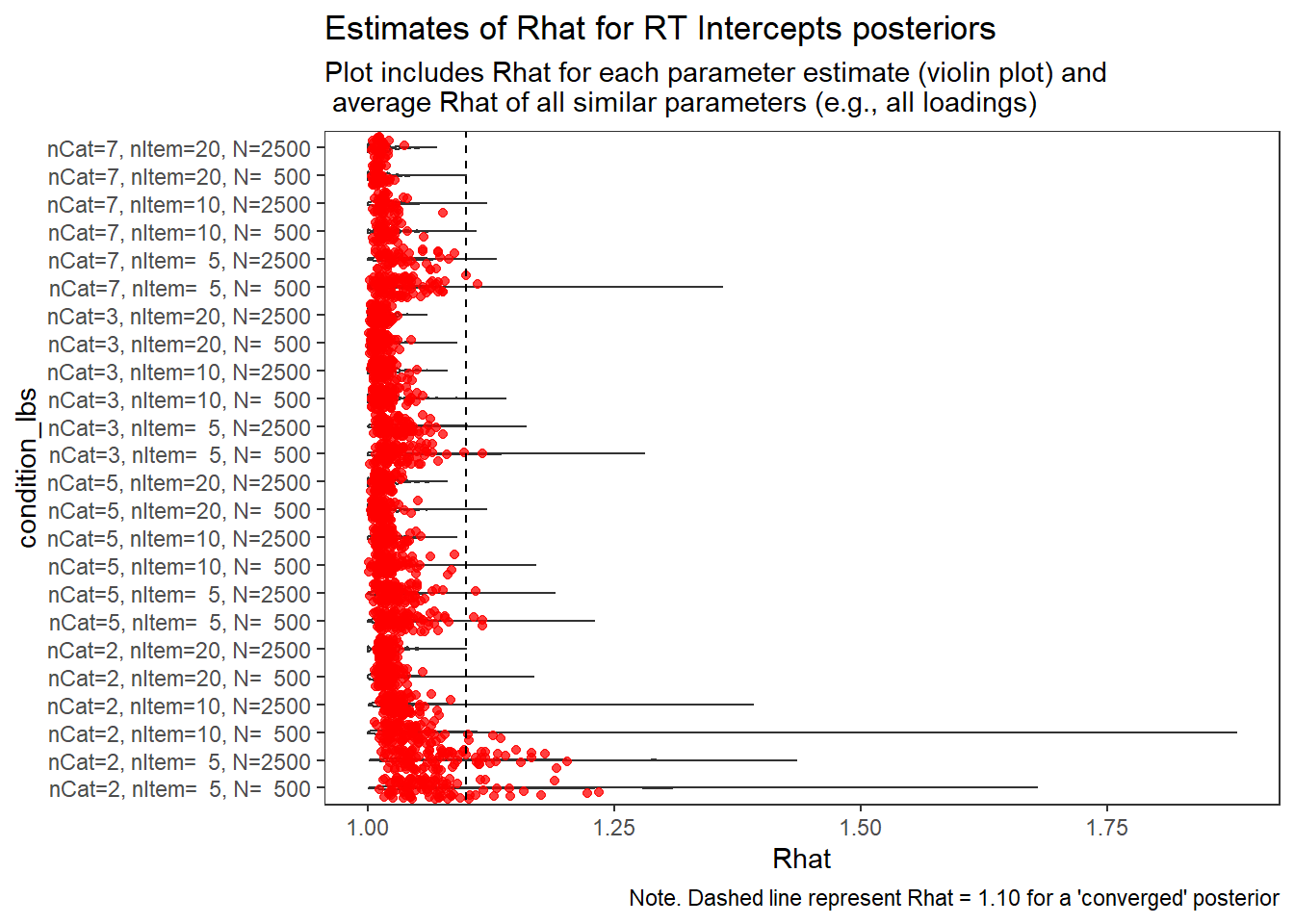
Response Time Intercepts (\(\beta_{lrt}\))
param = "RT Intercepts"
conv <- mydata %>%
filter(parameter_group == "beta_lrt")
conv_para_grp <- mydata %>%
filter(parameter_group == "beta_lrt") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 500 | 402 | 0.804 |
| 2 | 2 | 5 | 2500 | 500 | 401 | 0.802 |
| 3 | 2 | 10 | 500 | 1000 | 927 | 0.927 |
| 4 | 2 | 10 | 2500 | 1000 | 974 | 0.974 |
| 5 | 2 | 20 | 500 | 2000 | 1986 | 0.993 |
| 6 | 2 | 20 | 2500 | 2000 | 1999 | 1.000 |
| 7 | 5 | 5 | 500 | 500 | 464 | 0.928 |
| 8 | 5 | 5 | 2500 | 500 | 488 | 0.976 |
| 9 | 5 | 10 | 500 | 1000 | 984 | 0.984 |
| 10 | 5 | 10 | 2500 | 1000 | 1000 | 1.000 |
| 11 | 5 | 20 | 500 | 2000 | 1995 | 0.998 |
| 12 | 5 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 13 | 3 | 5 | 500 | 500 | 480 | 0.960 |
| 14 | 3 | 5 | 2500 | 500 | 494 | 0.988 |
| 15 | 3 | 10 | 500 | 1000 | 996 | 0.996 |
| 16 | 3 | 10 | 2500 | 1000 | 1000 | 1.000 |
| 17 | 3 | 20 | 500 | 2000 | 2000 | 1.000 |
| 18 | 3 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 19 | 7 | 5 | 500 | 500 | 482 | 0.964 |
| 20 | 7 | 5 | 2500 | 250 | 237 | 0.948 |
| 21 | 7 | 10 | 500 | 500 | 497 | 0.994 |
| 22 | 7 | 10 | 2500 | 500 | 497 | 0.994 |
| 23 | 7 | 20 | 500 | 1000 | 999 | 0.999 |
| 24 | 7 | 20 | 2500 | 760 | 760 | 1.000 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 85 | 0.85 |
| 2 | 2 | 5 | 2500 | 100 | 81 | 0.81 |
| 3 | 2 | 10 | 500 | 100 | 96 | 0.96 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1.00 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 97 | 0.97 |
| 8 | 5 | 5 | 2500 | 100 | 99 | 0.99 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 99 | 0.99 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 98 | 0.98 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1.00 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
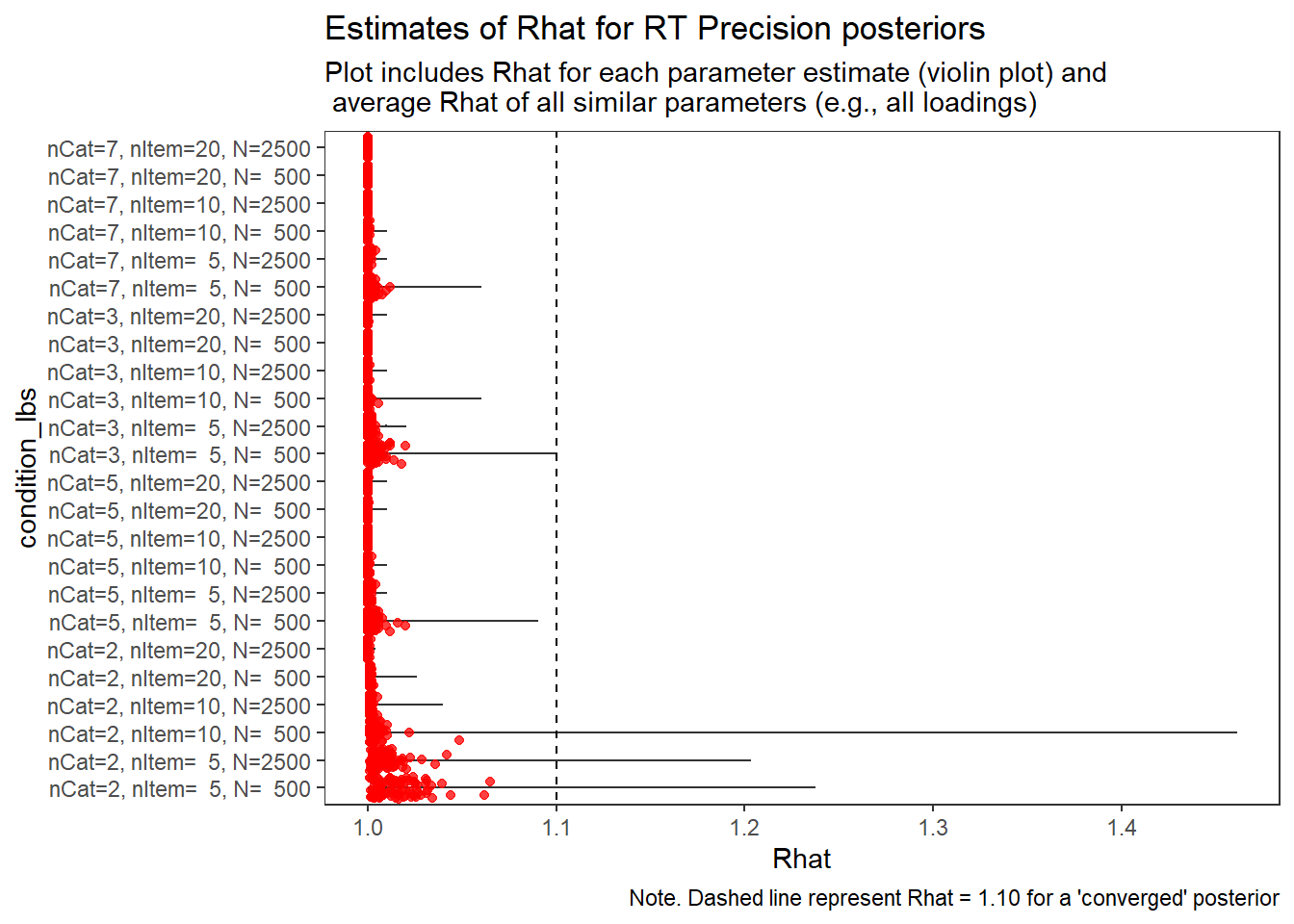
Response Time Precision (\(\sigma_{lrt}\))
param = "RT Precision"
conv <- mydata %>%
filter(parameter_group == "sigma_lrt")
conv_para_grp <- mydata %>%
filter(parameter_group == "sigma_lrt") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 500 | 485 | 0.970 |
| 2 | 2 | 5 | 2500 | 500 | 497 | 0.994 |
| 3 | 2 | 10 | 500 | 1000 | 998 | 0.998 |
| 4 | 2 | 10 | 2500 | 1000 | 1000 | 1.000 |
| 5 | 2 | 20 | 500 | 2000 | 2000 | 1.000 |
| 6 | 2 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 7 | 5 | 5 | 500 | 500 | 500 | 1.000 |
| 8 | 5 | 5 | 2500 | 500 | 500 | 1.000 |
| 9 | 5 | 10 | 500 | 1000 | 1000 | 1.000 |
| 10 | 5 | 10 | 2500 | 1000 | 1000 | 1.000 |
| 11 | 5 | 20 | 500 | 2000 | 2000 | 1.000 |
| 12 | 5 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 13 | 3 | 5 | 500 | 500 | 499 | 0.998 |
| 14 | 3 | 5 | 2500 | 500 | 500 | 1.000 |
| 15 | 3 | 10 | 500 | 1000 | 1000 | 1.000 |
| 16 | 3 | 10 | 2500 | 1000 | 1000 | 1.000 |
| 17 | 3 | 20 | 500 | 2000 | 2000 | 1.000 |
| 18 | 3 | 20 | 2500 | 2000 | 2000 | 1.000 |
| 19 | 7 | 5 | 500 | 500 | 500 | 1.000 |
| 20 | 7 | 5 | 2500 | 250 | 250 | 1.000 |
| 21 | 7 | 10 | 500 | 500 | 500 | 1.000 |
| 22 | 7 | 10 | 2500 | 500 | 500 | 1.000 |
| 23 | 7 | 20 | 500 | 1000 | 1000 | 1.000 |
| 24 | 7 | 20 | 2500 | 760 | 760 | 1.000 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 100 | 1 |
| 2 | 2 | 5 | 2500 | 100 | 100 | 1 |
| 3 | 2 | 10 | 500 | 100 | 100 | 1 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1 |
| 7 | 5 | 5 | 500 | 100 | 100 | 1 |
| 8 | 5 | 5 | 2500 | 100 | 100 | 1 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1 |
| 13 | 3 | 5 | 500 | 100 | 100 | 1 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1 |
| 19 | 7 | 5 | 500 | 100 | 100 | 1 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
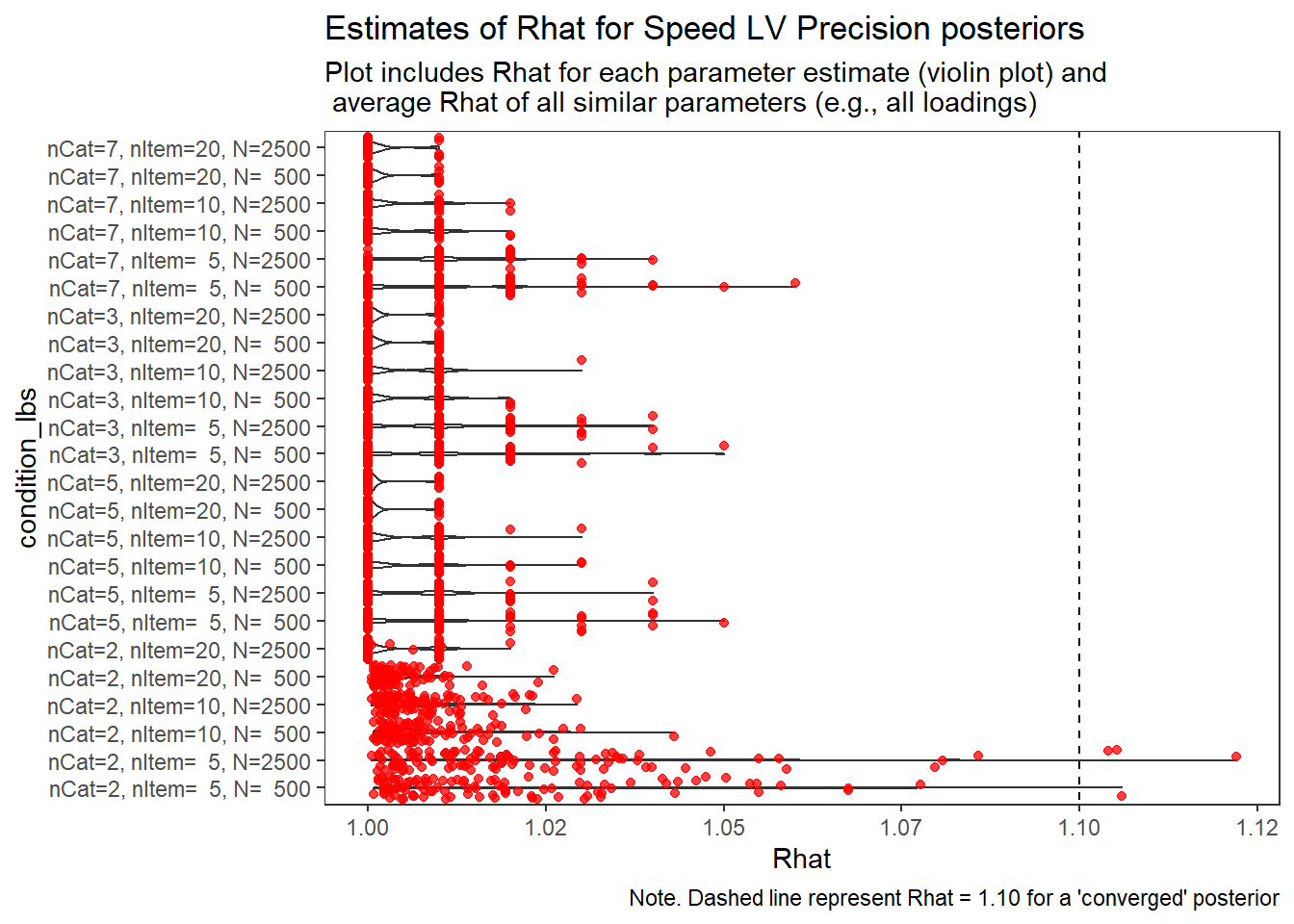
Response Time Factor Precision (\(\sigma_s\))
param = "Speed LV Precision"
conv <- mydata %>%
filter(parameter_group == "sigma_s")
conv_para_grp <- mydata %>%
filter(parameter_group == "sigma_s") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 99 | 0.99 |
| 2 | 2 | 5 | 2500 | 100 | 97 | 0.97 |
| 3 | 2 | 10 | 500 | 100 | 100 | 1.00 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1.00 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 100 | 1.00 |
| 8 | 5 | 5 | 2500 | 100 | 100 | 1.00 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 100 | 1.00 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 100 | 1.00 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1.00 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 99 | 0.99 |
| 2 | 2 | 5 | 2500 | 100 | 97 | 0.97 |
| 3 | 2 | 10 | 500 | 100 | 100 | 1.00 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1.00 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 100 | 1.00 |
| 8 | 5 | 5 | 2500 | 100 | 100 | 1.00 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 100 | 1.00 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 100 | 1.00 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1.00 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
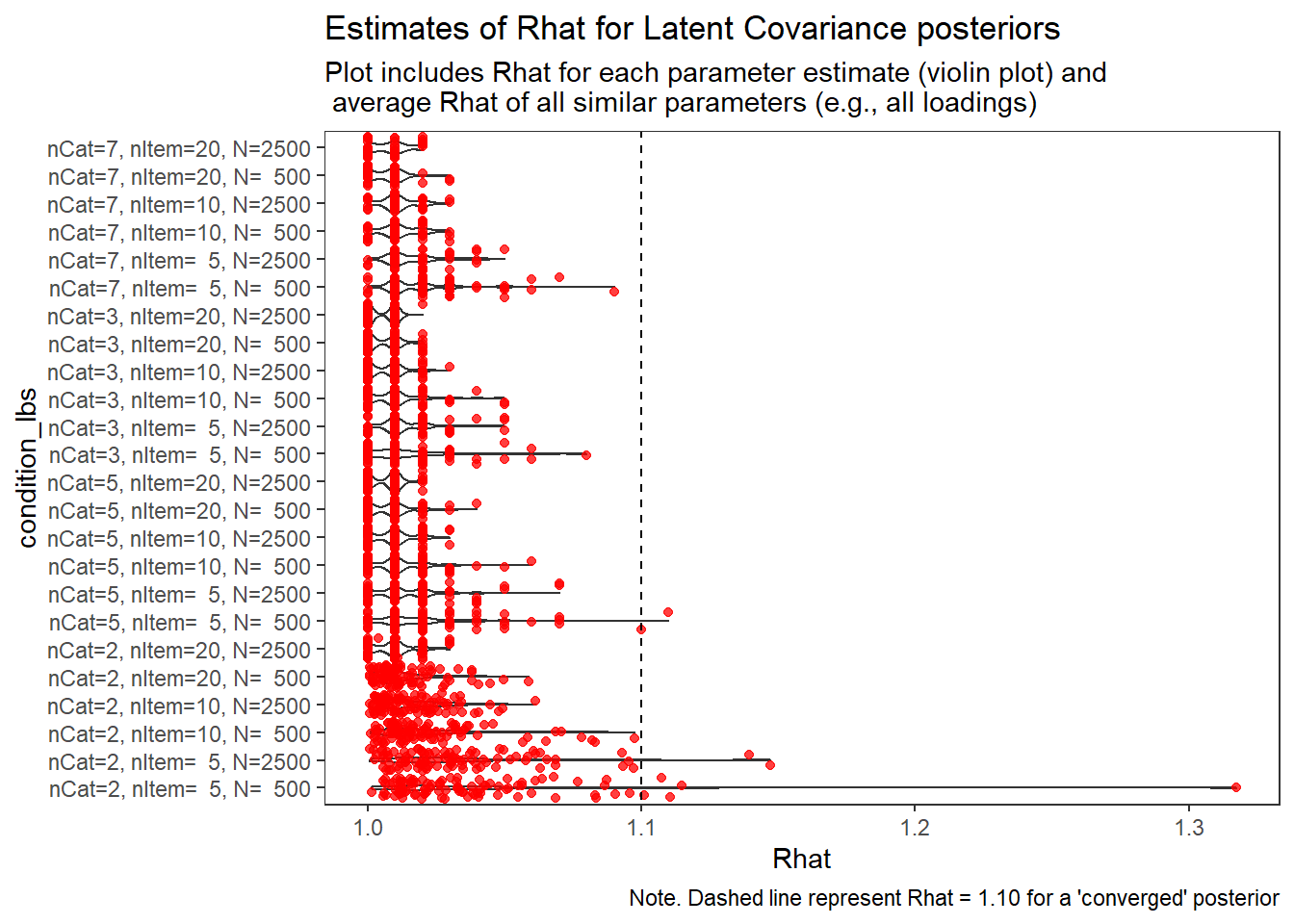
Factor Covariance (\(\sigma_{st}\))
param = "Latent Covariance"
conv <- mydata %>%
filter(parameter_group == "sigma_st")
conv_para_grp <- mydata %>%
filter(parameter_group == "sigma_st") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 95 | 0.95 |
| 2 | 2 | 5 | 2500 | 100 | 98 | 0.98 |
| 3 | 2 | 10 | 500 | 100 | 100 | 1.00 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1.00 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 98 | 0.98 |
| 8 | 5 | 5 | 2500 | 100 | 100 | 1.00 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 100 | 1.00 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 100 | 1.00 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1.00 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 95 | 0.95 |
| 2 | 2 | 5 | 2500 | 100 | 98 | 0.98 |
| 3 | 2 | 10 | 500 | 100 | 100 | 1.00 |
| 4 | 2 | 10 | 2500 | 100 | 100 | 1.00 |
| 5 | 2 | 20 | 500 | 100 | 100 | 1.00 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 98 | 0.98 |
| 8 | 5 | 5 | 2500 | 100 | 100 | 1.00 |
| 9 | 5 | 10 | 500 | 100 | 100 | 1.00 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 100 | 1.00 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 100 | 1.00 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 100 | 1.00 |
| 20 | 7 | 5 | 2500 | 50 | 50 | 1.00 |
| 21 | 7 | 10 | 500 | 50 | 50 | 1.00 |
| 22 | 7 | 10 | 2500 | 50 | 50 | 1.00 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
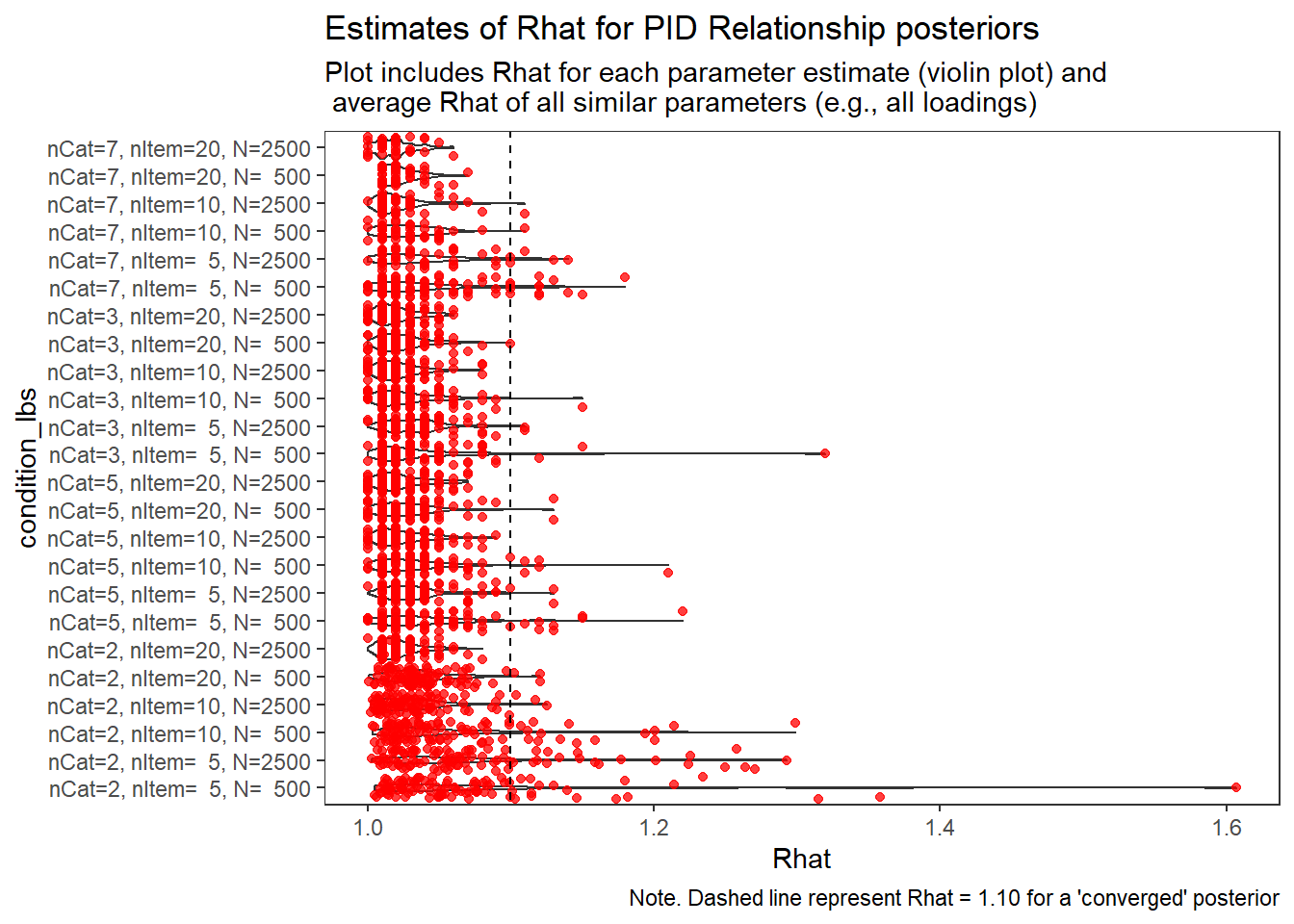
PID Relationship (\(\rho\))
param = "PID Relationship"
conv <- mydata %>%
filter(parameter_group == "rho")
conv_para_grp <- mydata %>%
filter(parameter_group == "rho") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 82 | 0.82 |
| 2 | 2 | 5 | 2500 | 100 | 80 | 0.80 |
| 3 | 2 | 10 | 500 | 100 | 85 | 0.85 |
| 4 | 2 | 10 | 2500 | 100 | 97 | 0.97 |
| 5 | 2 | 20 | 500 | 100 | 97 | 0.97 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 91 | 0.91 |
| 8 | 5 | 5 | 2500 | 100 | 97 | 0.97 |
| 9 | 5 | 10 | 500 | 100 | 94 | 0.94 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 98 | 0.98 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 97 | 0.97 |
| 14 | 3 | 5 | 2500 | 100 | 98 | 0.98 |
| 15 | 3 | 10 | 500 | 100 | 99 | 0.99 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 99 | 0.99 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 87 | 0.87 |
| 20 | 7 | 5 | 2500 | 50 | 45 | 0.90 |
| 21 | 7 | 10 | 500 | 50 | 49 | 0.98 |
| 22 | 7 | 10 | 2500 | 50 | 49 | 0.98 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 82 | 0.82 |
| 2 | 2 | 5 | 2500 | 100 | 80 | 0.80 |
| 3 | 2 | 10 | 500 | 100 | 85 | 0.85 |
| 4 | 2 | 10 | 2500 | 100 | 97 | 0.97 |
| 5 | 2 | 20 | 500 | 100 | 97 | 0.97 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 91 | 0.91 |
| 8 | 5 | 5 | 2500 | 100 | 97 | 0.97 |
| 9 | 5 | 10 | 500 | 100 | 94 | 0.94 |
| 10 | 5 | 10 | 2500 | 100 | 100 | 1.00 |
| 11 | 5 | 20 | 500 | 100 | 98 | 0.98 |
| 12 | 5 | 20 | 2500 | 100 | 100 | 1.00 |
| 13 | 3 | 5 | 500 | 100 | 97 | 0.97 |
| 14 | 3 | 5 | 2500 | 100 | 98 | 0.98 |
| 15 | 3 | 10 | 500 | 100 | 99 | 0.99 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 99 | 0.99 |
| 18 | 3 | 20 | 2500 | 100 | 100 | 1.00 |
| 19 | 7 | 5 | 500 | 100 | 87 | 0.87 |
| 20 | 7 | 5 | 2500 | 50 | 45 | 0.90 |
| 21 | 7 | 10 | 500 | 50 | 49 | 0.98 |
| 22 | 7 | 10 | 2500 | 50 | 49 | 0.98 |
| 23 | 7 | 20 | 500 | 50 | 50 | 1.00 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
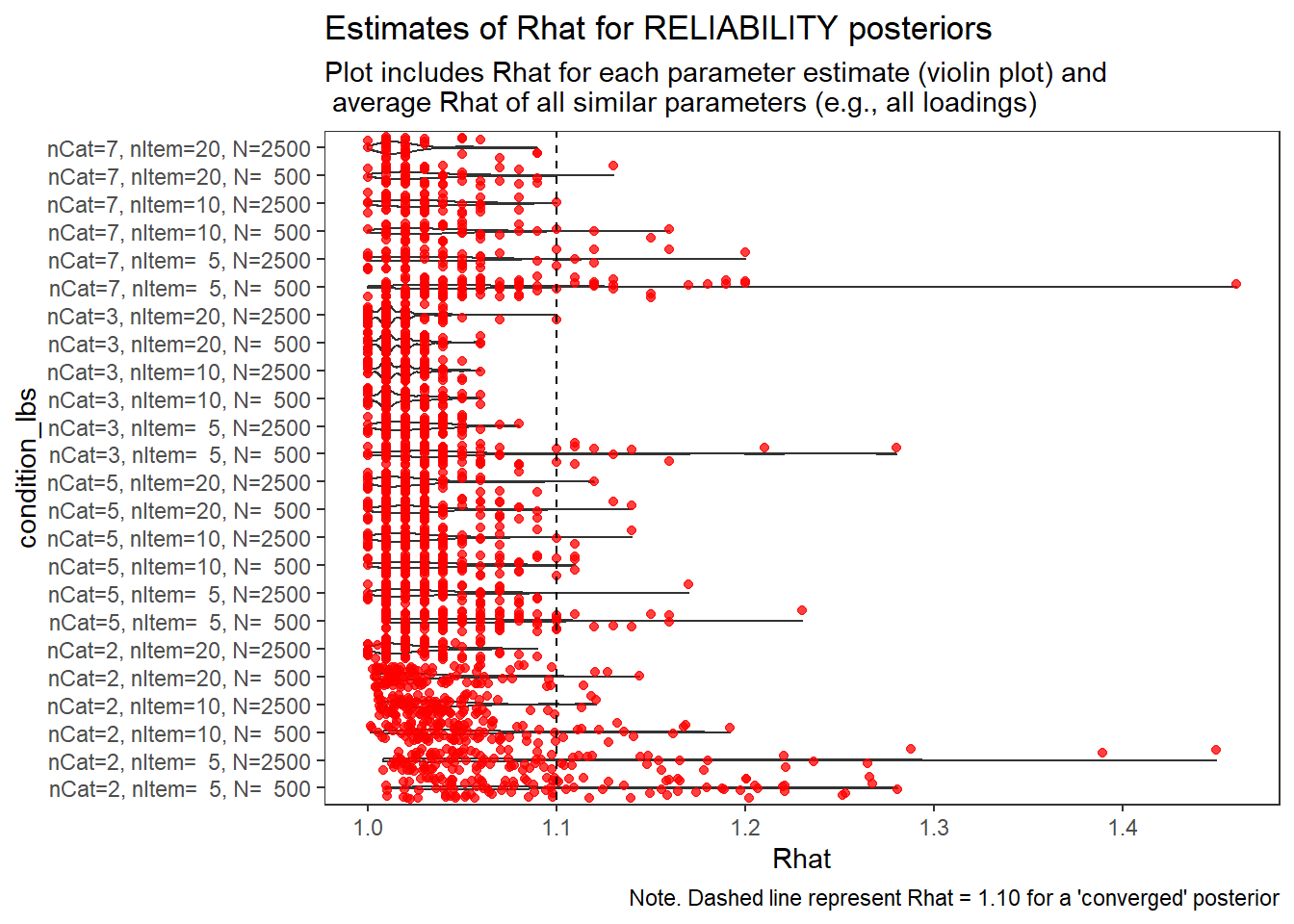
Factor Reliability (\(\omega\))
param = "RELIABILITY"
conv <- mydata %>%
filter(parameter_group == "omega")
conv_para_grp <- mydata %>%
filter(parameter_group == "omega") %>%
select(all_of(keep_cols)) %>%
distinct()
# tabular summary
conv_tbl <- conv %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 63 | 0.63 |
| 2 | 2 | 5 | 2500 | 100 | 78 | 0.78 |
| 3 | 2 | 10 | 500 | 100 | 88 | 0.88 |
| 4 | 2 | 10 | 2500 | 100 | 97 | 0.97 |
| 5 | 2 | 20 | 500 | 100 | 96 | 0.96 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 87 | 0.87 |
| 8 | 5 | 5 | 2500 | 100 | 99 | 0.99 |
| 9 | 5 | 10 | 500 | 100 | 95 | 0.95 |
| 10 | 5 | 10 | 2500 | 100 | 97 | 0.97 |
| 11 | 5 | 20 | 500 | 100 | 98 | 0.98 |
| 12 | 5 | 20 | 2500 | 100 | 99 | 0.99 |
| 13 | 3 | 5 | 500 | 100 | 89 | 0.89 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 99 | 0.99 |
| 19 | 7 | 5 | 500 | 100 | 81 | 0.81 |
| 20 | 7 | 5 | 2500 | 50 | 43 | 0.86 |
| 21 | 7 | 10 | 500 | 50 | 46 | 0.92 |
| 22 | 7 | 10 | 2500 | 50 | 49 | 0.98 |
| 23 | 7 | 20 | 500 | 50 | 49 | 0.98 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
conv_grp_tbl <- conv_para_grp %>%
group_by(condition, N_cat, N_items, N) %>%
summarise(Max_Converged=n(),
Obs_Converged=sum(converge_paragrp),
Prop_Converged=Obs_Converged/Max_Converged)`summarise()` has grouped output by 'condition', 'N_cat', 'N_items'. You can override using the `.groups` argument.kable(conv_grp_tbl, format="html", digits=3) %>%
kable_styling(full_width = T)| condition | N_cat | N_items | N | Max_Converged | Obs_Converged | Prop_Converged |
|---|---|---|---|---|---|---|
| 1 | 2 | 5 | 500 | 100 | 63 | 0.63 |
| 2 | 2 | 5 | 2500 | 100 | 78 | 0.78 |
| 3 | 2 | 10 | 500 | 100 | 88 | 0.88 |
| 4 | 2 | 10 | 2500 | 100 | 97 | 0.97 |
| 5 | 2 | 20 | 500 | 100 | 96 | 0.96 |
| 6 | 2 | 20 | 2500 | 100 | 100 | 1.00 |
| 7 | 5 | 5 | 500 | 100 | 87 | 0.87 |
| 8 | 5 | 5 | 2500 | 100 | 99 | 0.99 |
| 9 | 5 | 10 | 500 | 100 | 95 | 0.95 |
| 10 | 5 | 10 | 2500 | 100 | 97 | 0.97 |
| 11 | 5 | 20 | 500 | 100 | 98 | 0.98 |
| 12 | 5 | 20 | 2500 | 100 | 99 | 0.99 |
| 13 | 3 | 5 | 500 | 100 | 89 | 0.89 |
| 14 | 3 | 5 | 2500 | 100 | 100 | 1.00 |
| 15 | 3 | 10 | 500 | 100 | 100 | 1.00 |
| 16 | 3 | 10 | 2500 | 100 | 100 | 1.00 |
| 17 | 3 | 20 | 500 | 100 | 100 | 1.00 |
| 18 | 3 | 20 | 2500 | 100 | 99 | 0.99 |
| 19 | 7 | 5 | 500 | 100 | 81 | 0.81 |
| 20 | 7 | 5 | 2500 | 50 | 43 | 0.86 |
| 21 | 7 | 10 | 500 | 50 | 46 | 0.92 |
| 22 | 7 | 10 | 2500 | 50 | 49 | 0.98 |
| 23 | 7 | 20 | 500 | 50 | 49 | 0.98 |
| 24 | 7 | 20 | 2500 | 38 | 38 | 1.00 |
# visualize Rhat values
conv_grp <- conv %>%
select(condition_lbs, iter, N_cat, N_items, N, Rhat_paragrp, converge_paragrp) %>%
distinct()Adding missing grouping variables: `condition`, `parameter_group`ggplot() +
geom_violin(data=conv, aes(x=condition_lbs , y=Rhat, group=condition))+
geom_jitter(data=conv_grp, aes(x=condition_lbs , y=Rhat_paragrp), color="red", alpha=0.75)+
geom_abline(intercept = 1.10, slope=0, linetype="dashed")+
coord_flip()+
labs(
title=paste0("Estimates of Rhat for ", param, " posteriors" ),
subtitle ="Plot includes Rhat for each parameter estimate (violin plot) and\n average Rhat of all similar parameters (e.g., all loadings)",
caption = "Note. Dashed line represent Rhat = 1.10 for a 'converged' posterior"
) +
theme_bw()+
theme(
panel.grid = element_blank()
)
Effect of Simulated Conditions
Factor Loadings (\(\lambda\))
conv_para_grp <- mydata %>%
filter(parameter_group == "lambda") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.8, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 79.9 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 241 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 10.3 0.0014 **
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.375 0.1249 269.23 < 0.0000000000000002 ***
N_items 2 0.440 0.2202 474.59 < 0.0000000000000002 ***
N 1 0.042 0.0416 89.65 < 0.0000000000000002 ***
N_cat:N_items 6 0.107 0.0179 38.48 < 0.0000000000000002 ***
N_cat:N 3 0.005 0.0015 3.31 0.019 *
N_items:N 2 0.003 0.0015 3.15 0.043 *
N_cat:N_items:N 6 0.027 0.0045 9.75 0.00000000013 ***
Residuals 2114 0.981 0.0005
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.1886 0.2735 0.18933
N_items 0.2220 0.3070 0.22249
N 0.0208 0.0398 0.02101
N_cat:N_items 0.0527 0.0952 0.05412
N_cat:N 0.0016 0.0032 0.00232
N_items:N 0.0010 0.0020 0.00148
N_cat:N_items:N 0.0123 0.0240 0.01371saved_omega[1,] <- omega2(fit)
saved_eta[1,] <- eta_estItem Thresholds (\(\tau\))
conv_para_grp <- mydata %>%
filter(parameter_group == "tau") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality: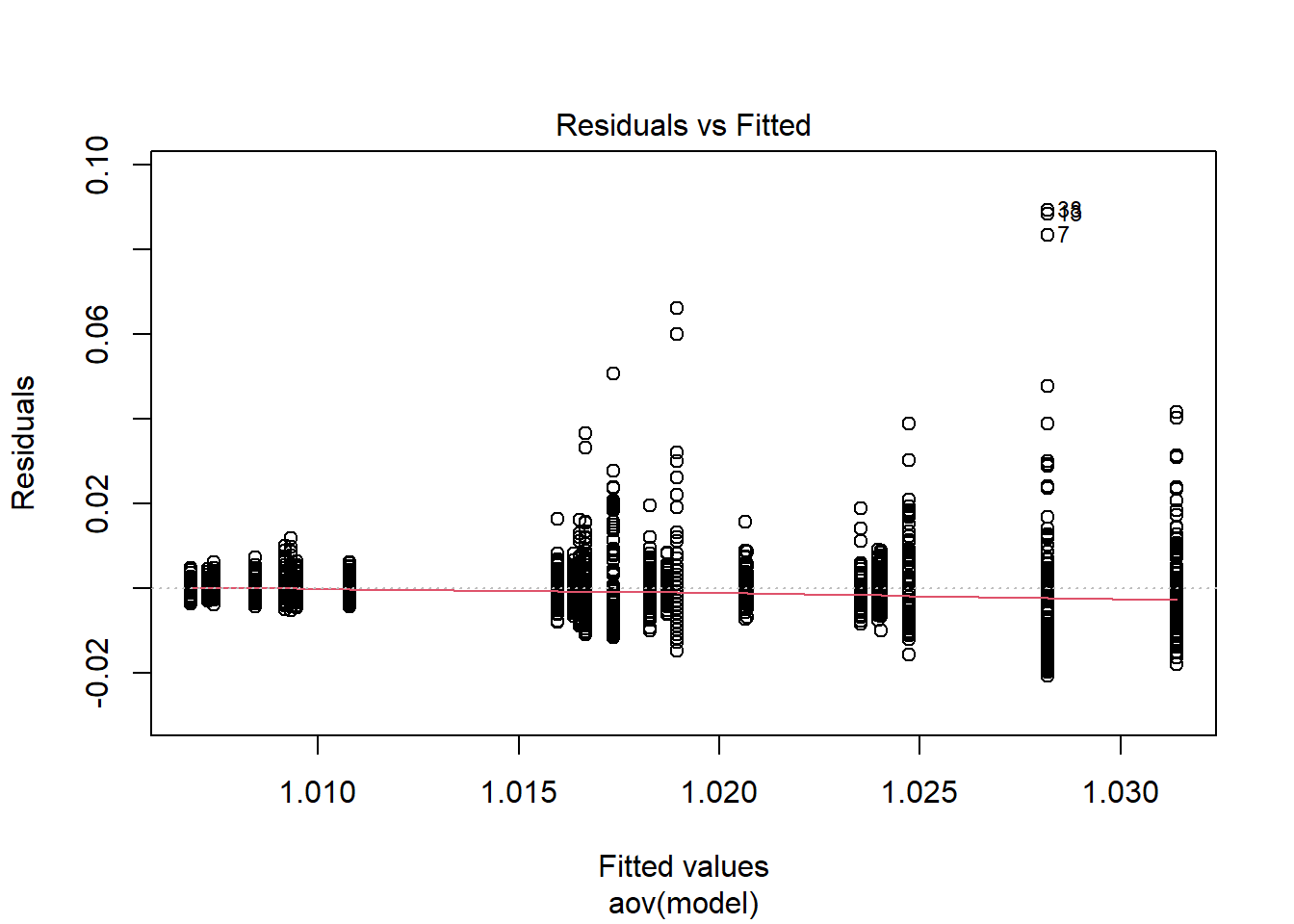
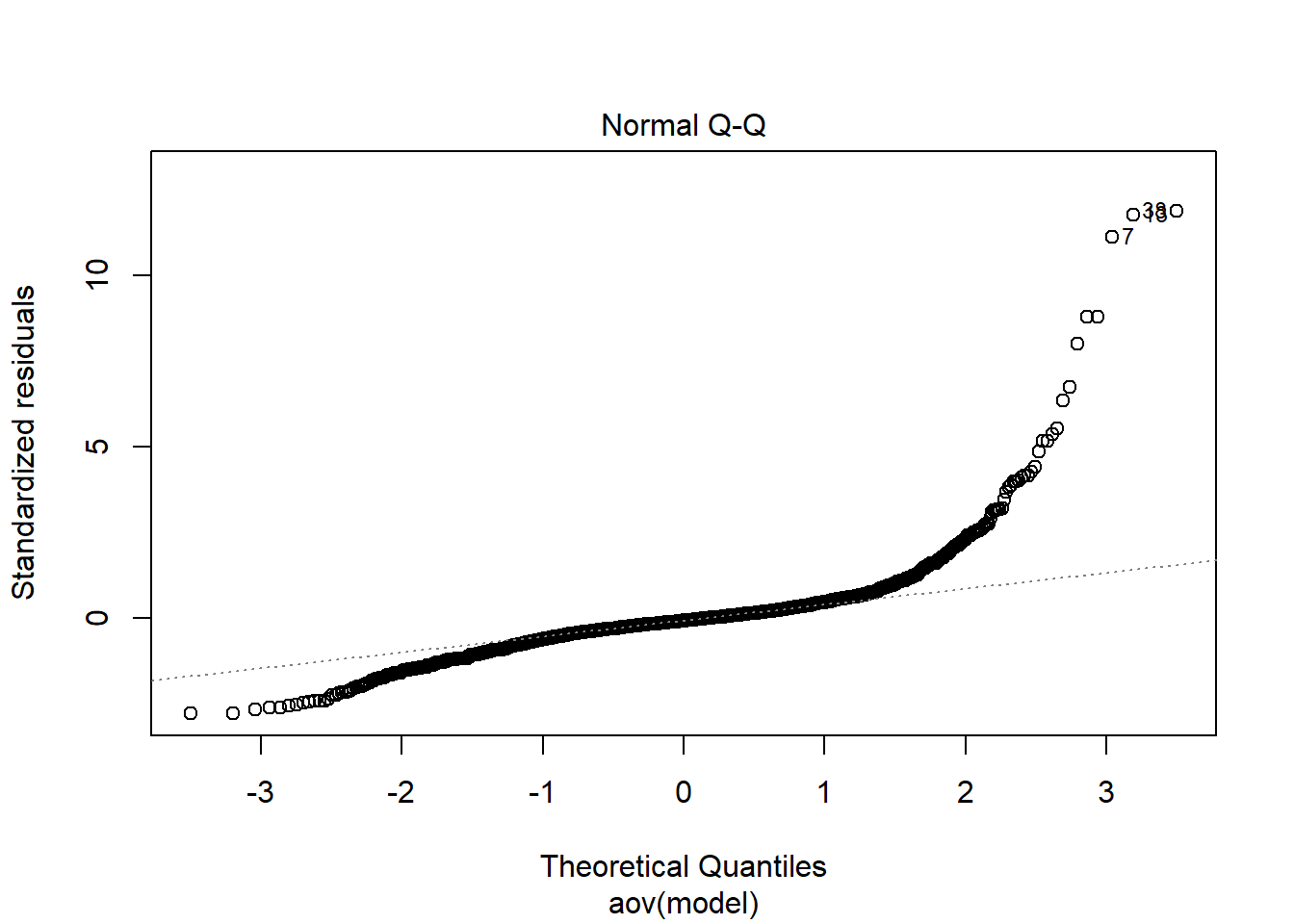
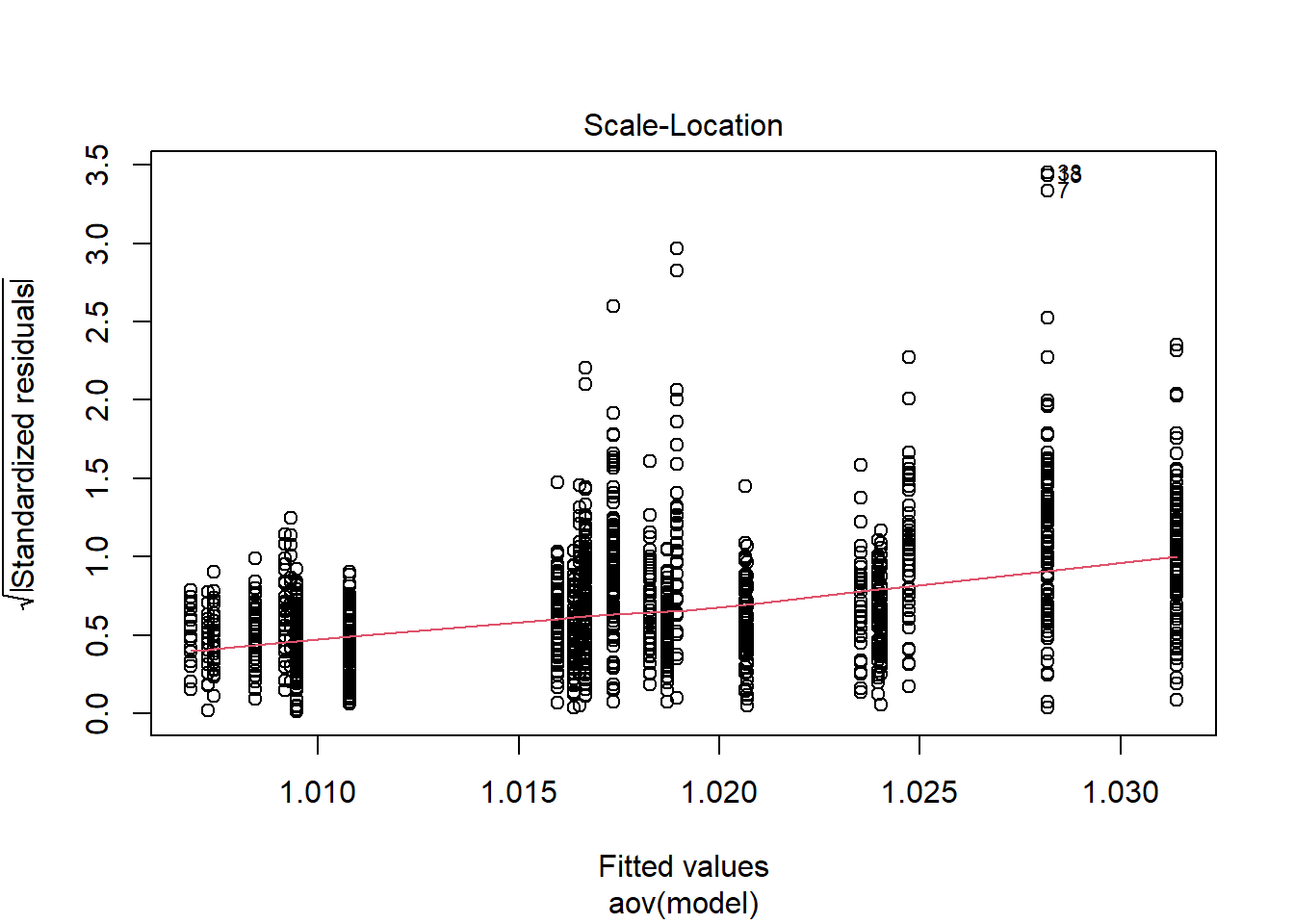
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.7, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test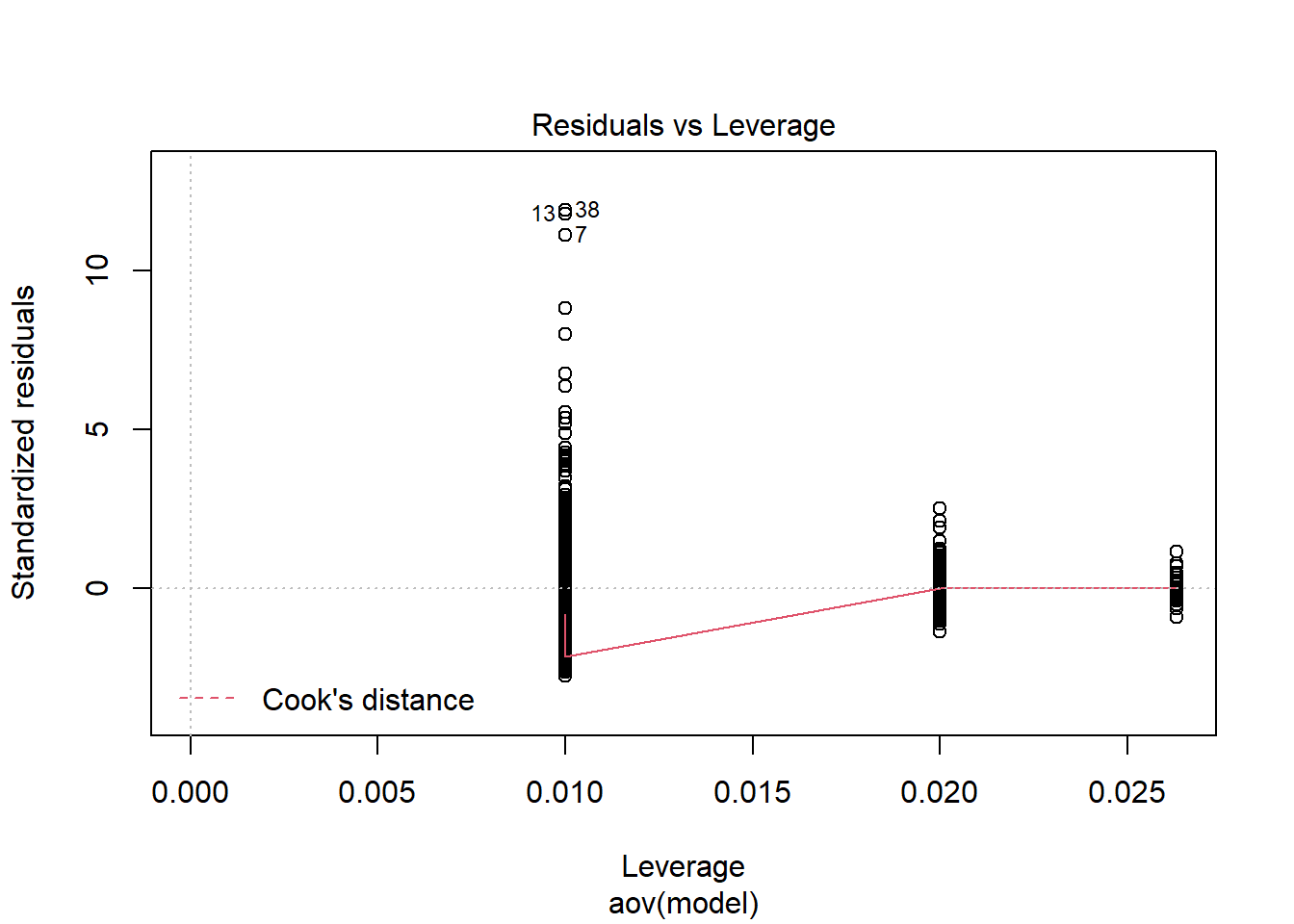
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.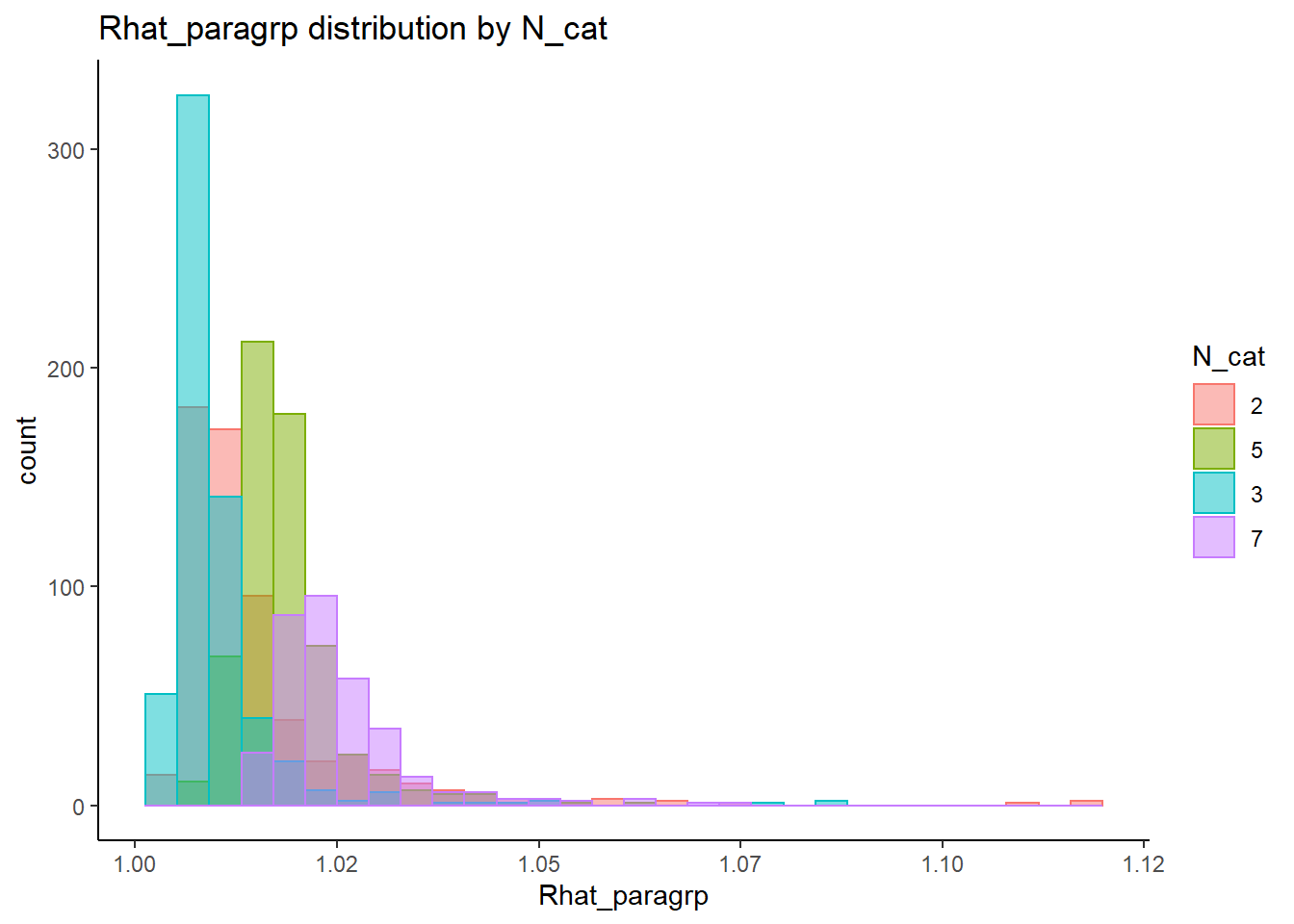
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.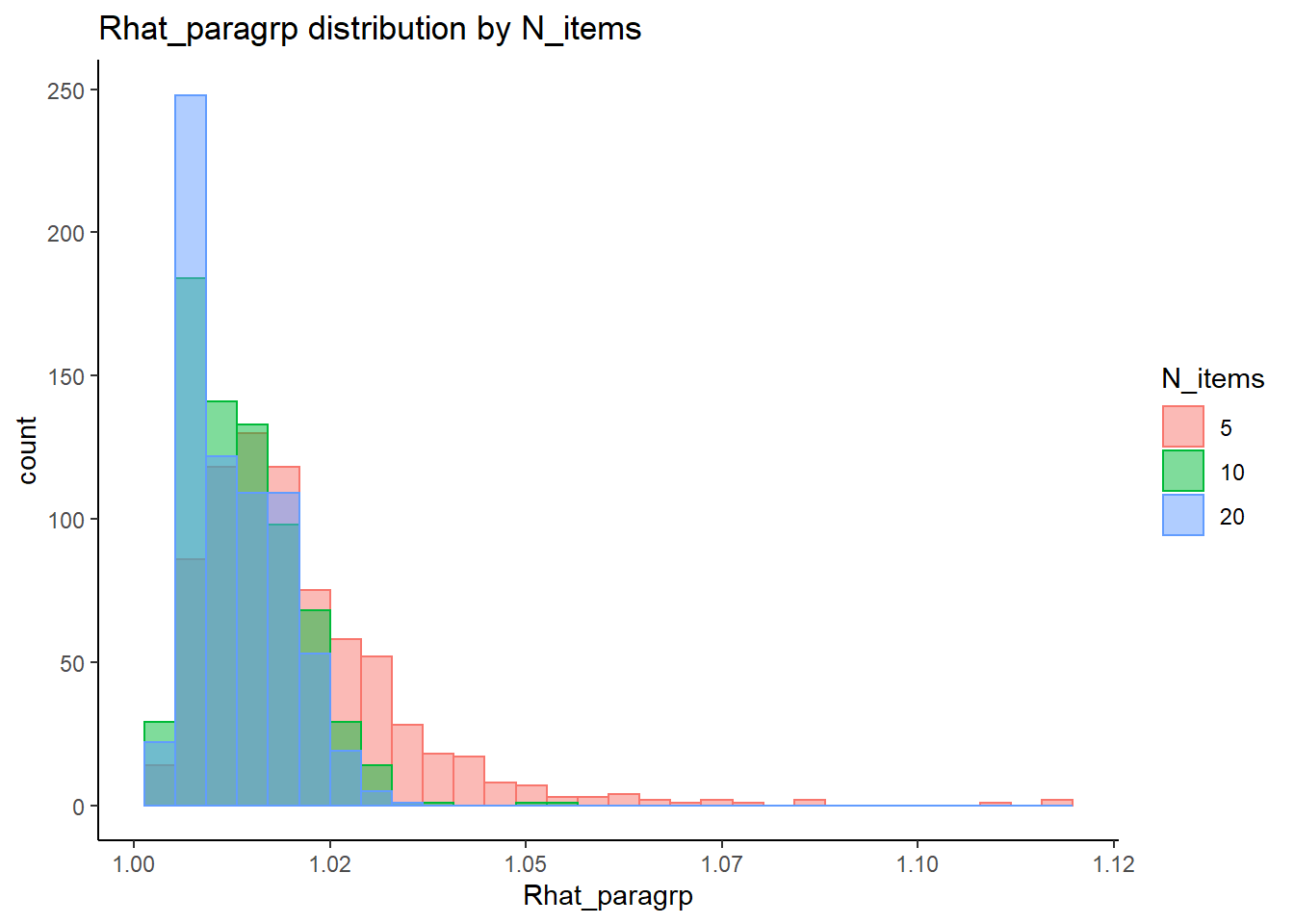
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.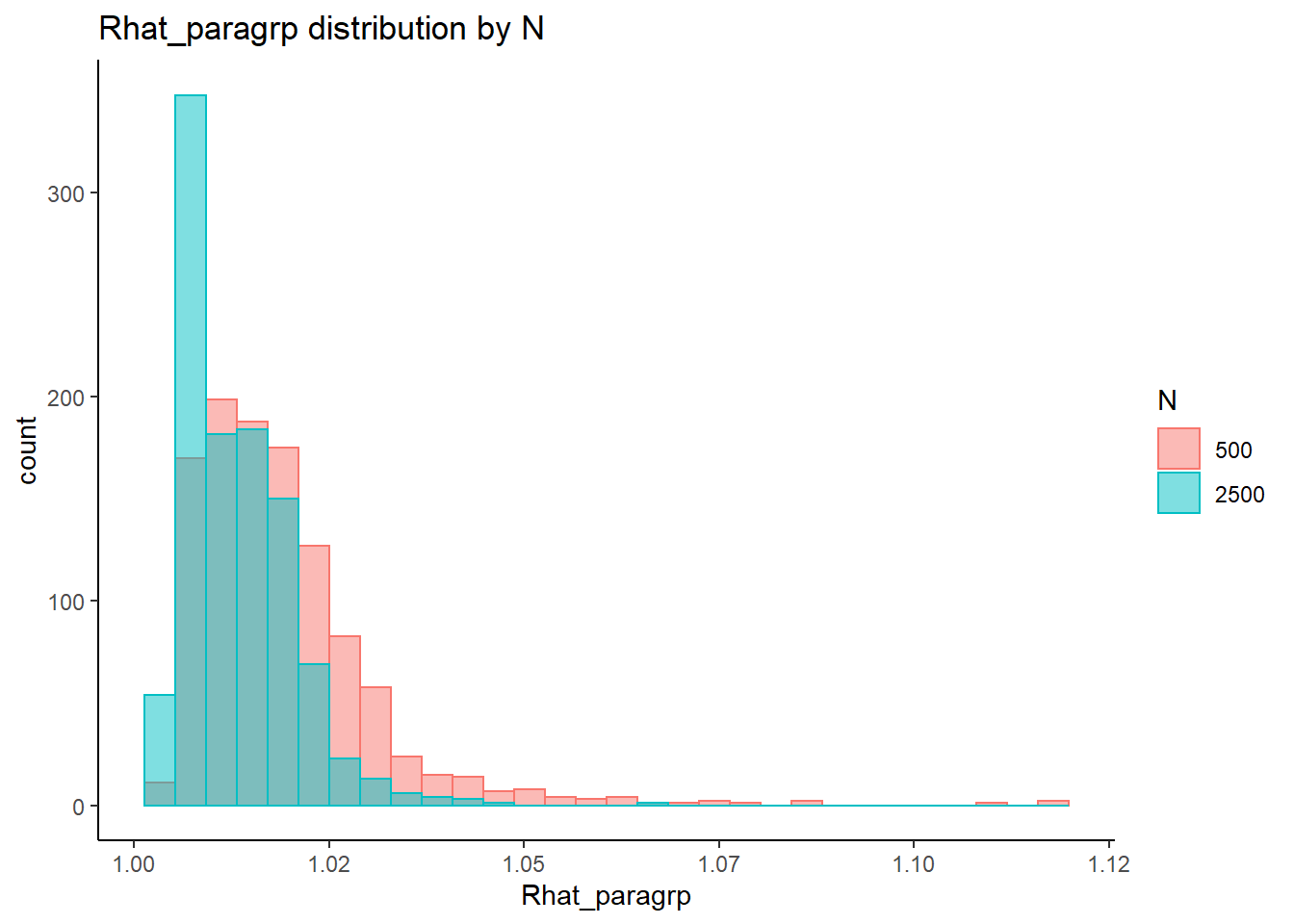
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 32.3 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 93.2 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 76 <0.0000000000000002 ***
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.0547 0.01824 319.94 <0.0000000000000002 ***
N_items 2 0.0237 0.01183 207.48 <0.0000000000000002 ***
N 1 0.0146 0.01459 255.89 <0.0000000000000002 ***
N_cat:N_items 6 0.0062 0.00103 18.05 <0.0000000000000002 ***
N_cat:N 3 0.0008 0.00026 4.62 0.0032 **
N_items:N 2 0.0047 0.00233 40.90 <0.0000000000000002 ***
N_cat:N_items:N 6 0.0004 0.00006 1.10 0.3580
Residuals 2114 0.1205 0.00006
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.2419 0.3092 0.24267
N_items 0.1044 0.1619 0.10492
N 0.0644 0.1065 0.06470
N_cat:N_items 0.0259 0.0457 0.02738
N_cat:N 0.0027 0.0050 0.00350
N_items:N 0.0202 0.0360 0.02068
N_cat:N_items:N 0.0002 0.0003 0.00167saved_omega[2,] <- omega2(fit)
saved_eta[2,] <- eta_estLatent Response Variance (\(\theta\))
conv_para_grp <- mydata %>%
filter(parameter_group == "theta") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality: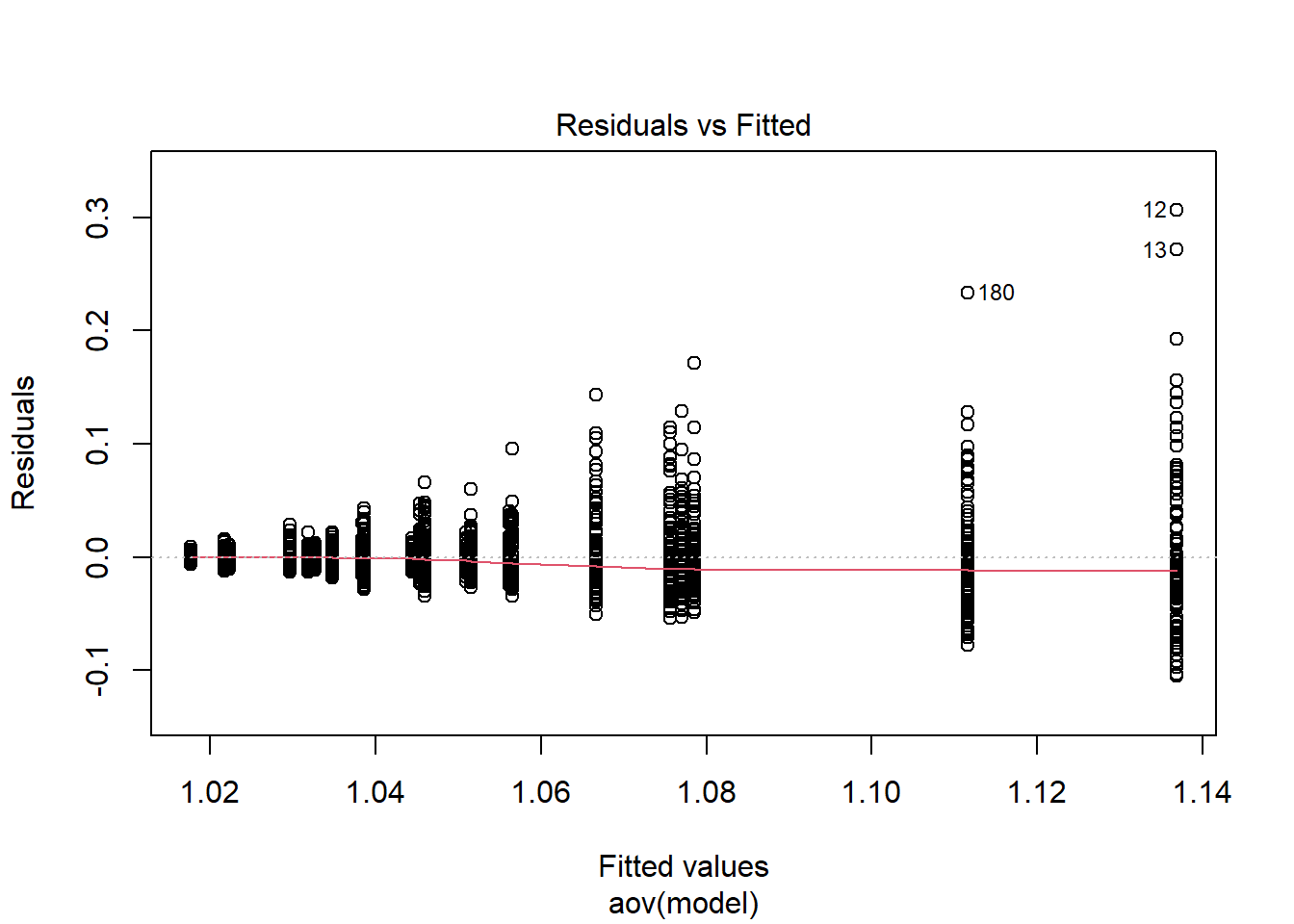
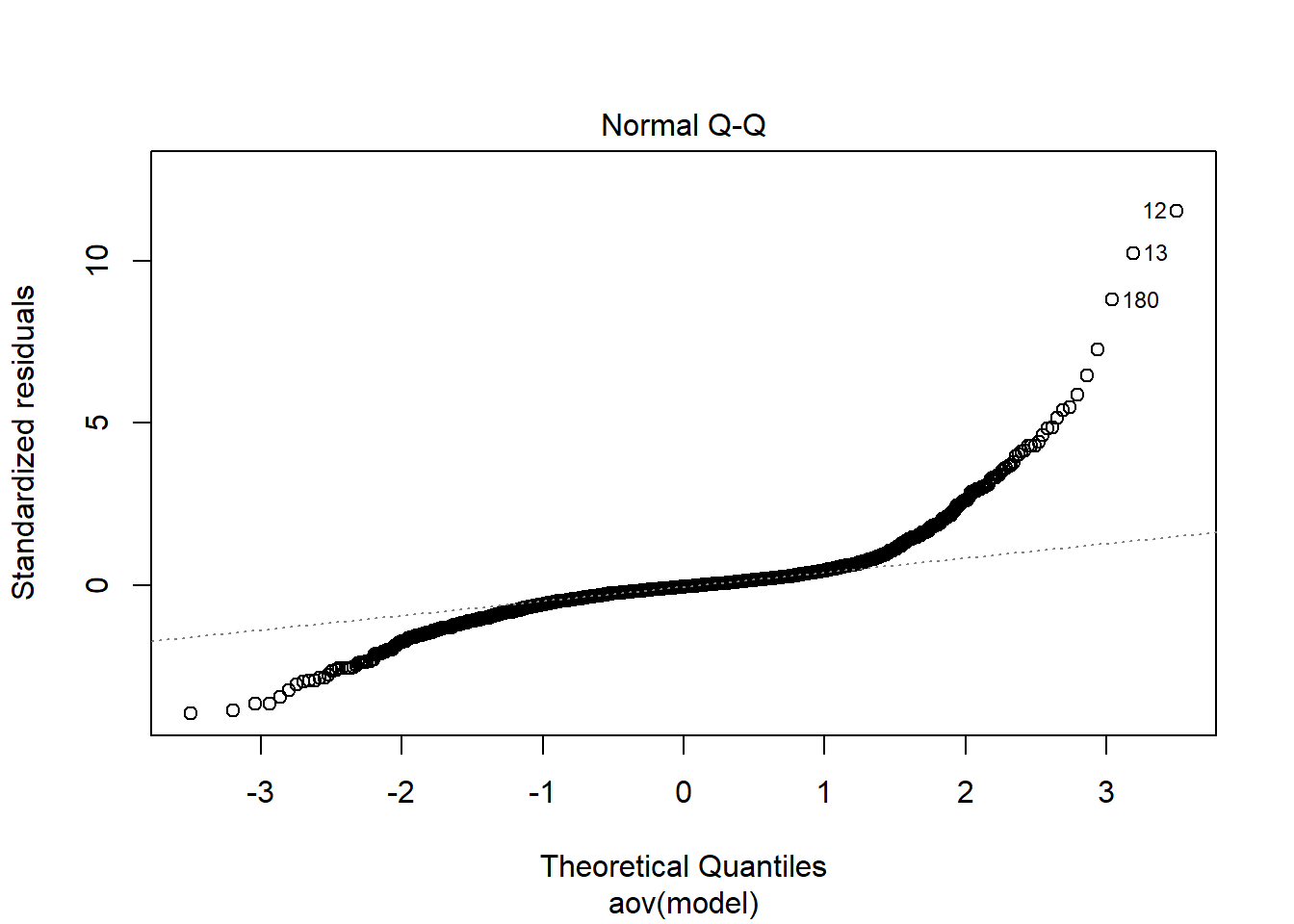
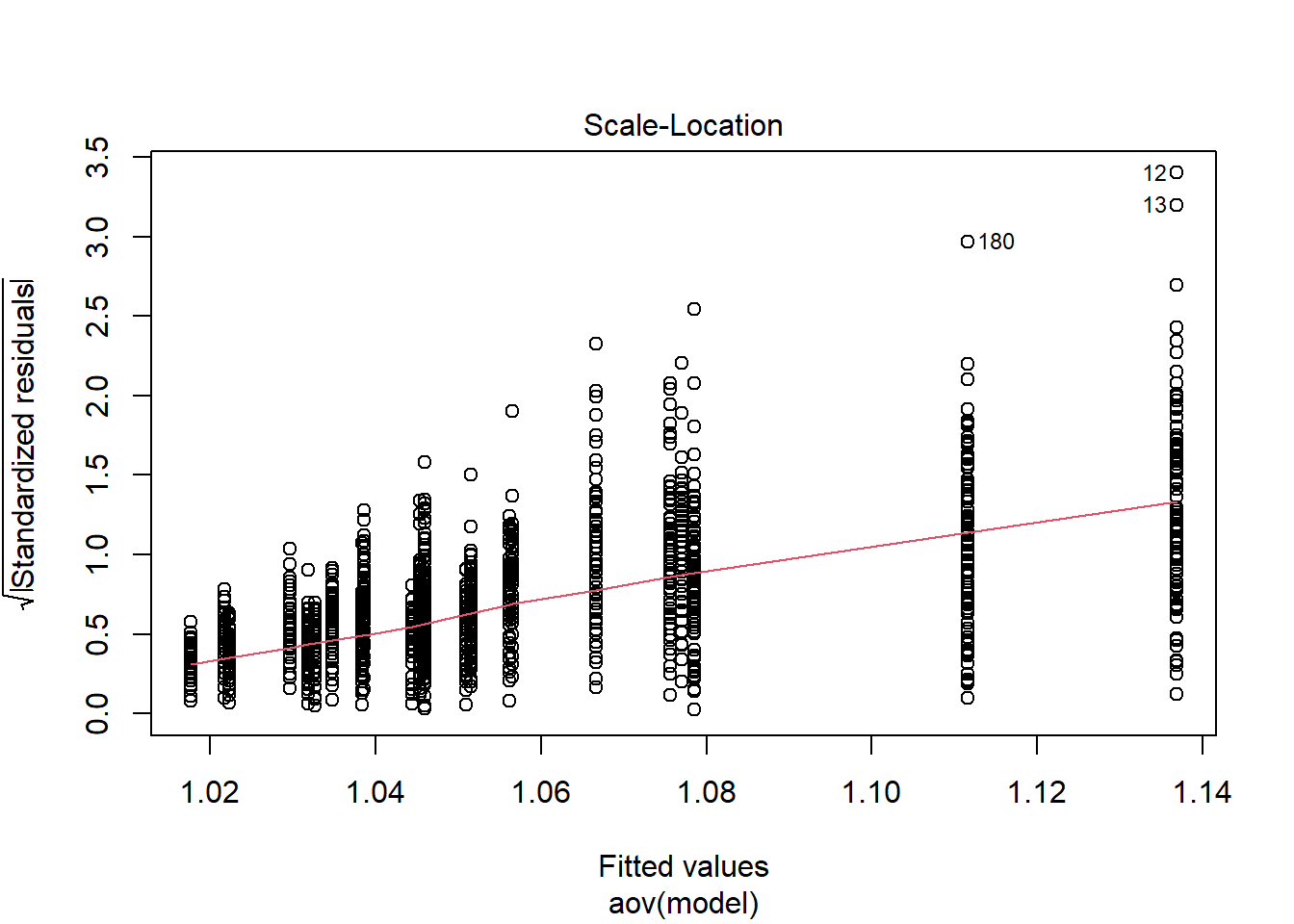
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.8, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.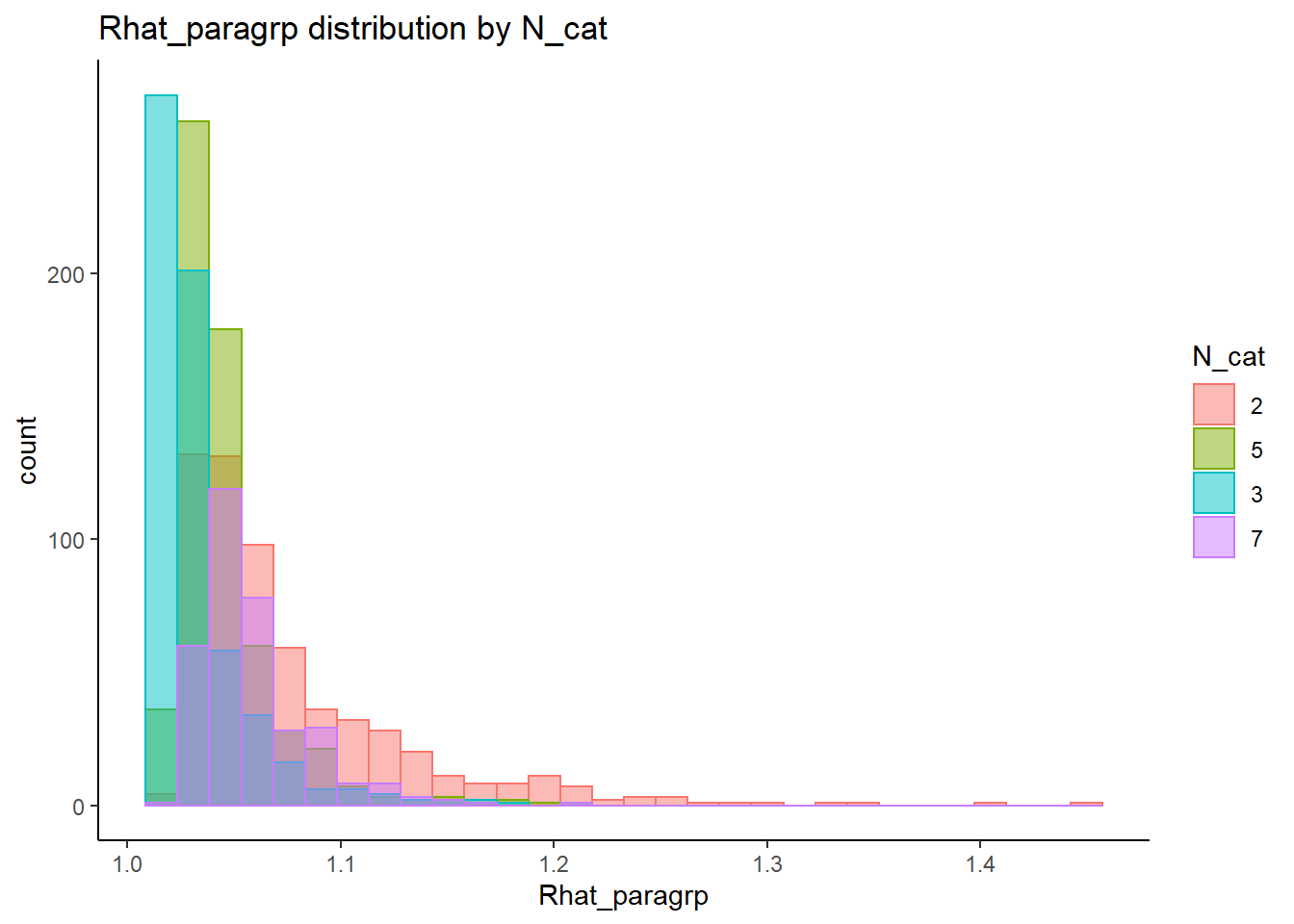
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.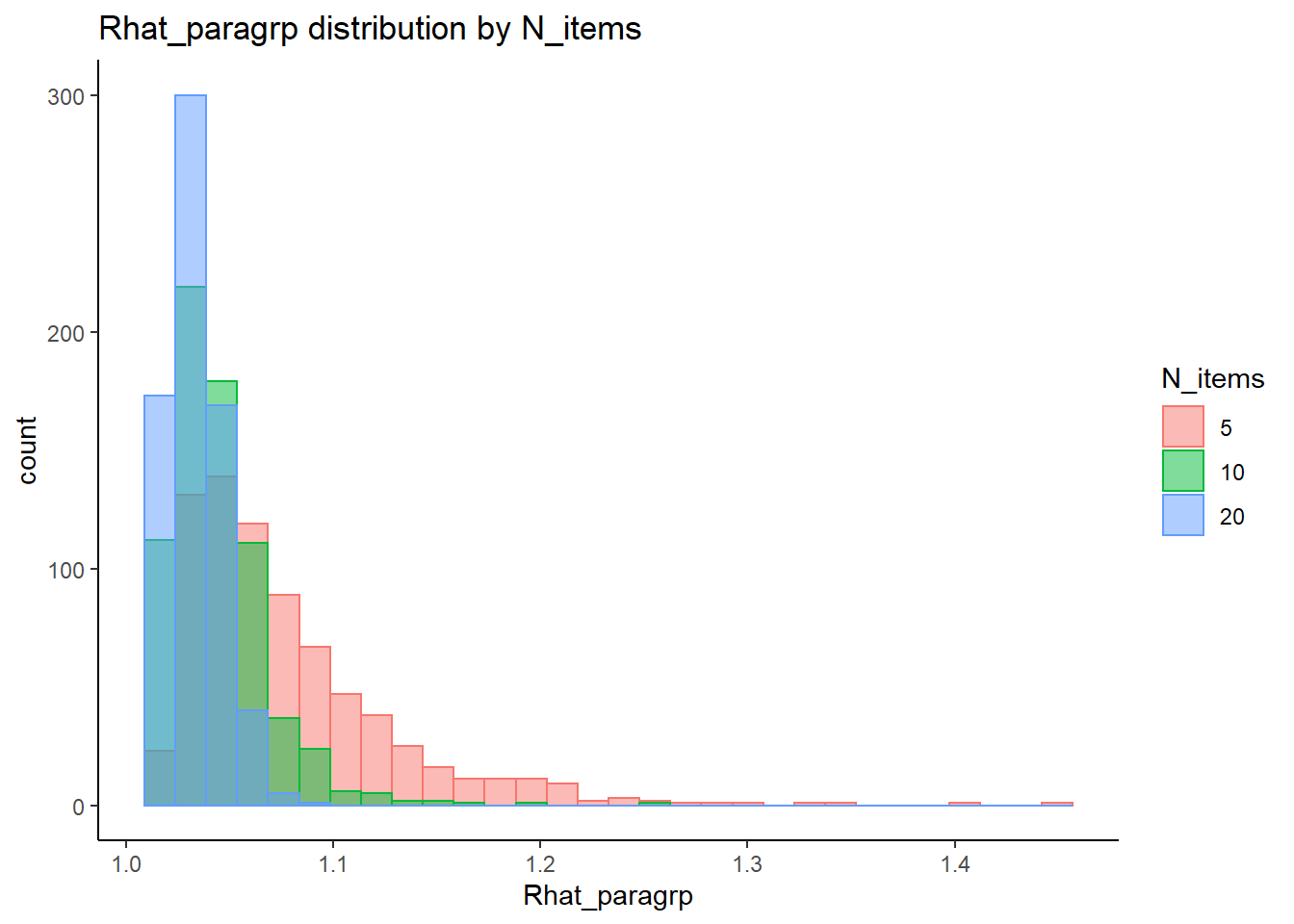
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.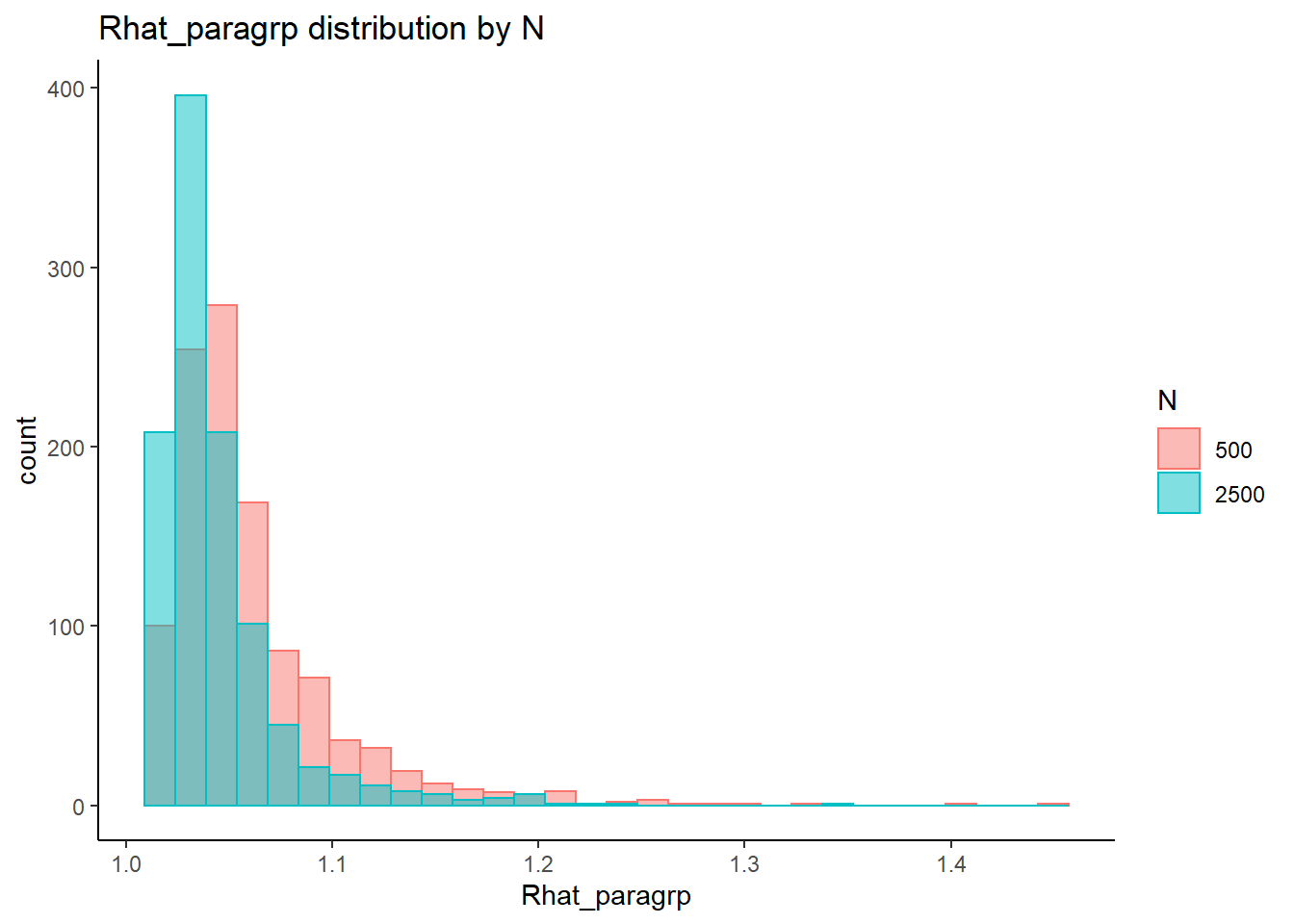
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 124 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 258 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 54.8 0.00000000000019 ***
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.614 0.205 288.25 < 0.0000000000000002 ***
N_items 2 0.741 0.371 522.07 < 0.0000000000000002 ***
N 1 0.138 0.138 194.78 < 0.0000000000000002 ***
N_cat:N_items 6 0.254 0.042 59.55 < 0.0000000000000002 ***
N_cat:N 3 0.006 0.002 2.96 0.031 *
N_items:N 2 0.032 0.016 22.37 0.00000000024 ***
N_cat:N_items:N 6 0.008 0.001 1.88 0.080 .
Residuals 2114 1.501 0.001
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.1857 0.2873 0.18637
N_items 0.2246 0.3277 0.22504
N 0.0418 0.0831 0.04198
N_cat:N_items 0.0757 0.1411 0.07700
N_cat:N 0.0013 0.0027 0.00192
N_items:N 0.0092 0.0196 0.00964
N_cat:N_items:N 0.0011 0.0025 0.00243saved_omega[3,] <- omega2(fit)
saved_eta[3,] <- eta_estResponse Time Intercepts (\(\beta_{lrt}\))
conv_para_grp <- mydata %>%
filter(parameter_group == "beta_lrt") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
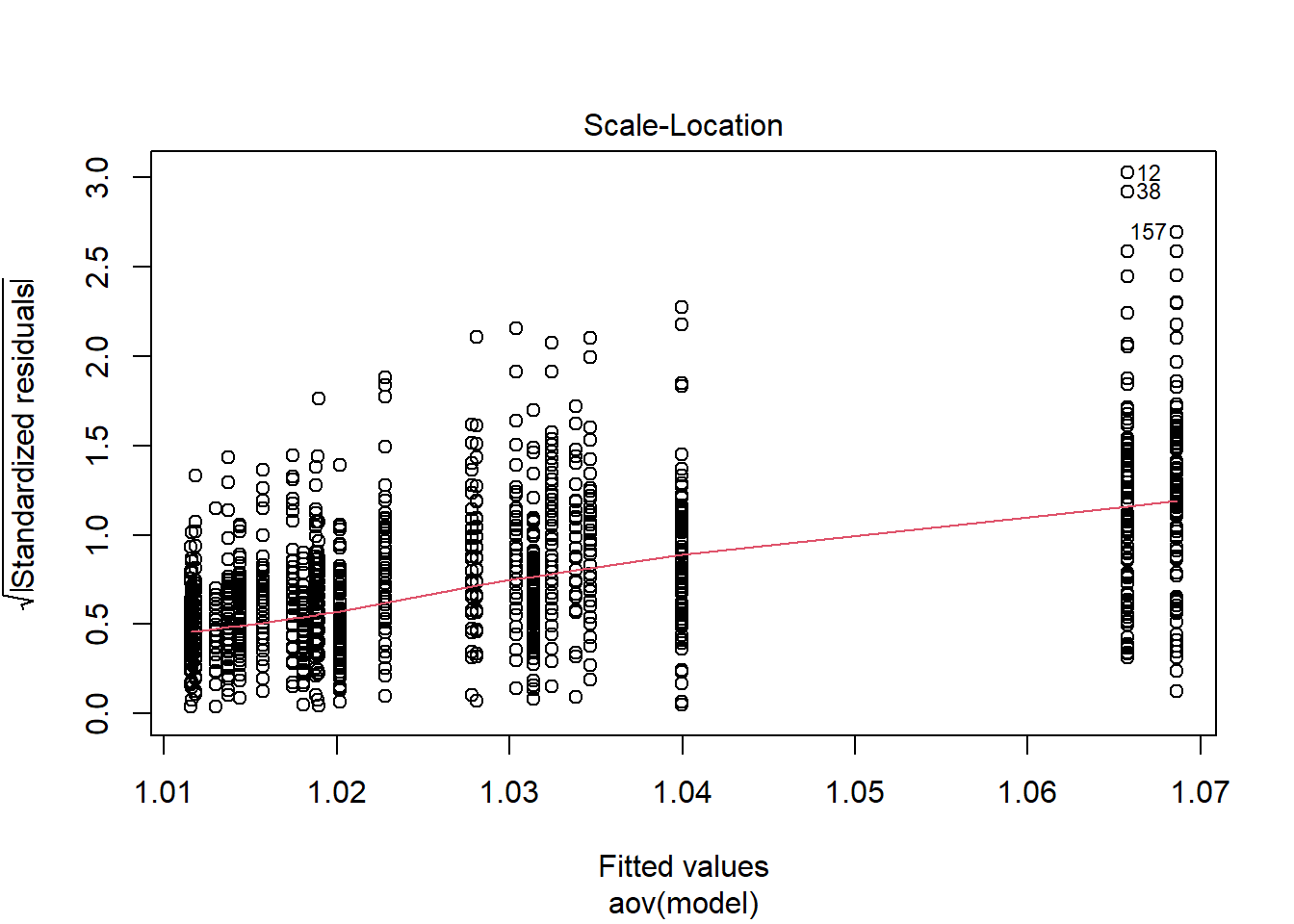
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.8, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.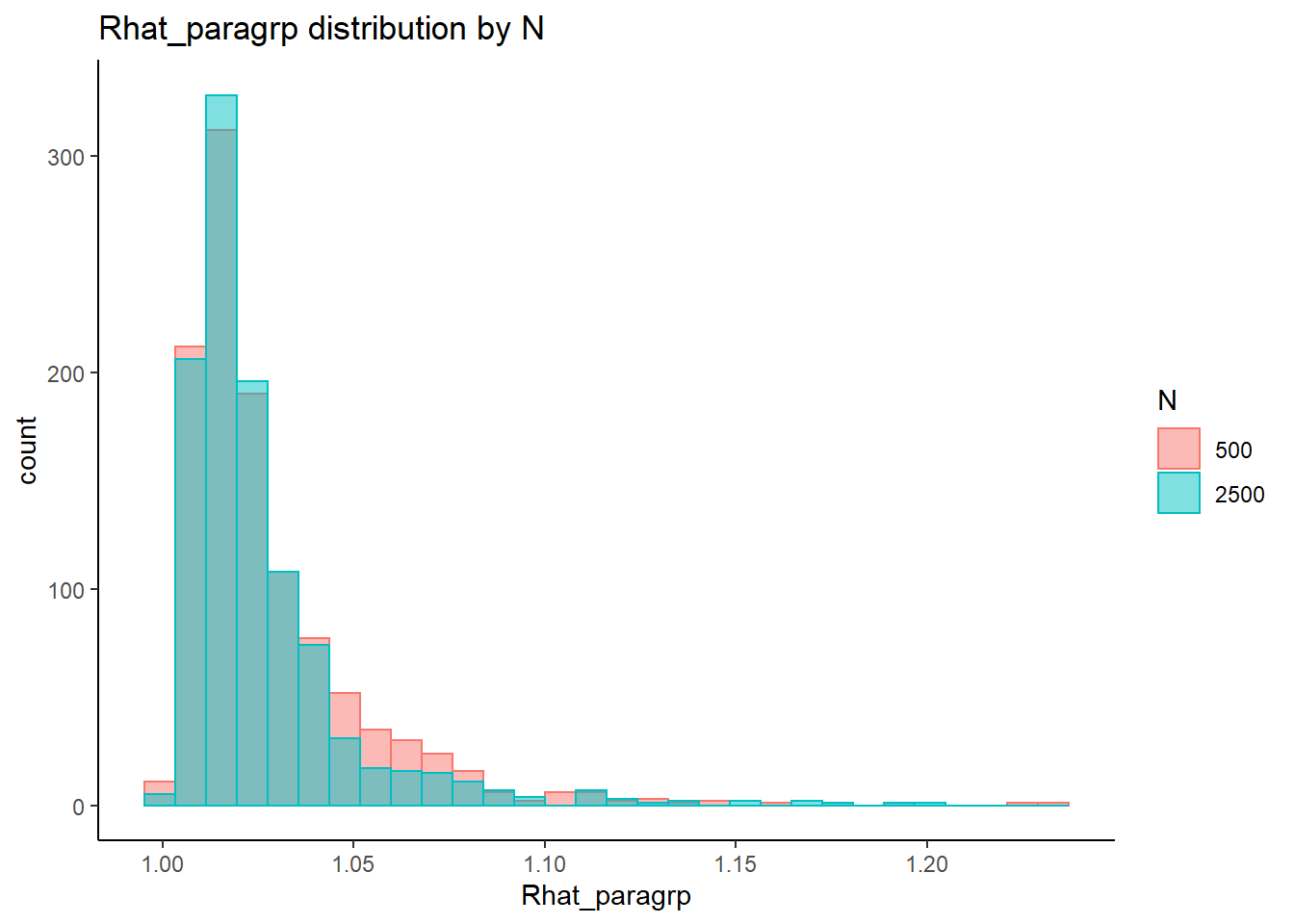
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 86.4 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 233 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 8.15 0.0043 **
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.168 0.0560 163.03 <0.0000000000000002 ***
N_items 2 0.261 0.1303 379.56 <0.0000000000000002 ***
N 1 0.002 0.0018 5.35 0.021 *
N_cat:N_items 6 0.068 0.0113 32.80 <0.0000000000000002 ***
N_cat:N 3 0.001 0.0004 1.04 0.375
N_items:N 2 0.001 0.0007 1.94 0.145
N_cat:N_items:N 6 0.003 0.0006 1.69 0.118
Residuals 2114 0.726 0.0003
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.1357 0.1852 0.136558
N_items 0.2113 0.2615 0.211958
N 0.0012 0.0020 0.001493
N_cat:N_items 0.0533 0.0819 0.054942
N_cat:N 0.0000 0.0001 0.000868
N_items:N 0.0005 0.0009 0.001081
N_cat:N_items:N 0.0012 0.0019 0.002839saved_omega[4,] <- omega2(fit)
saved_eta[4,] <- eta_estResponse Time Precision (\(\sigma_{lrt}\))
conv_para_grp <- mydata %>%
filter(parameter_group == "sigma_lrt") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.5, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test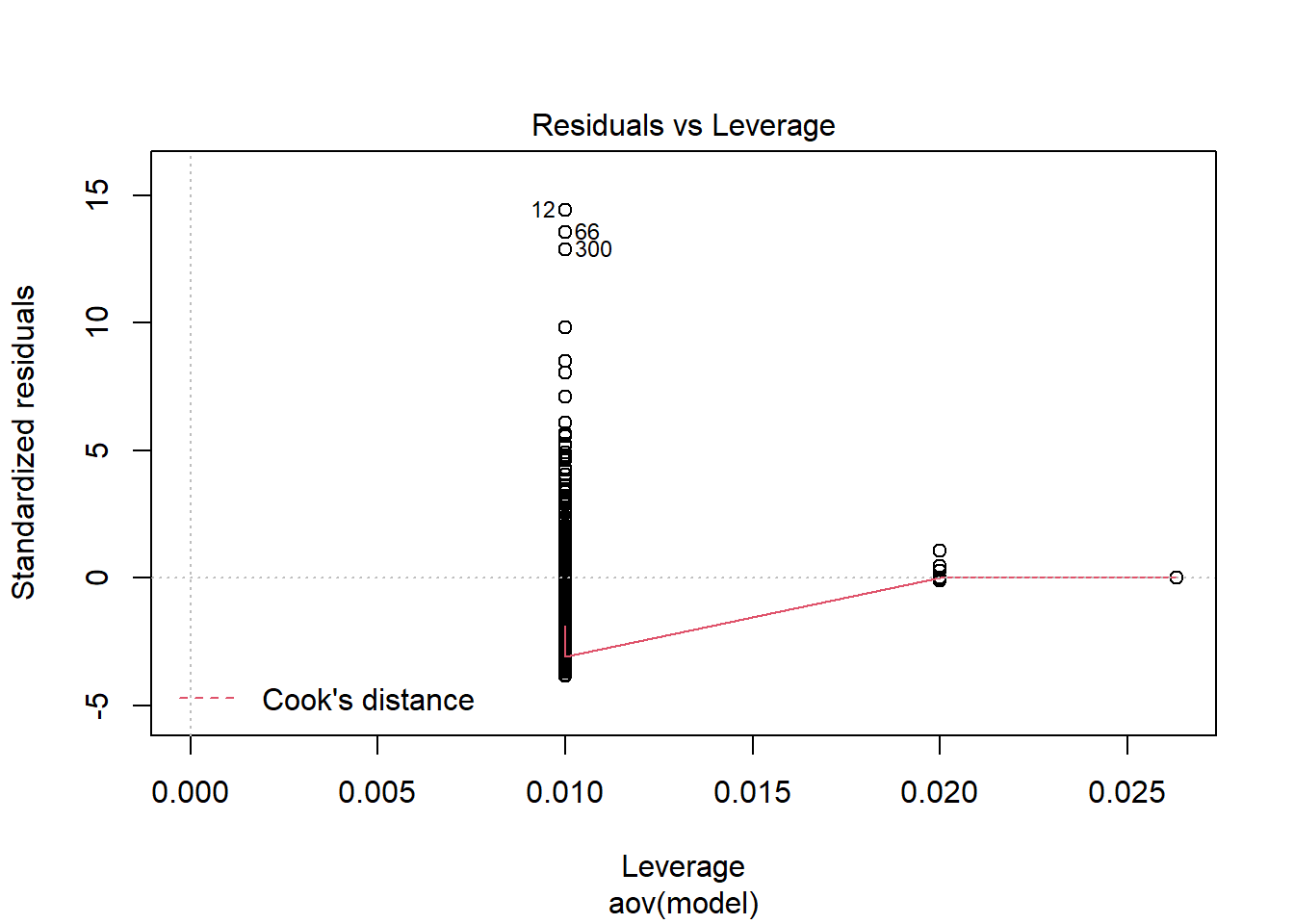
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.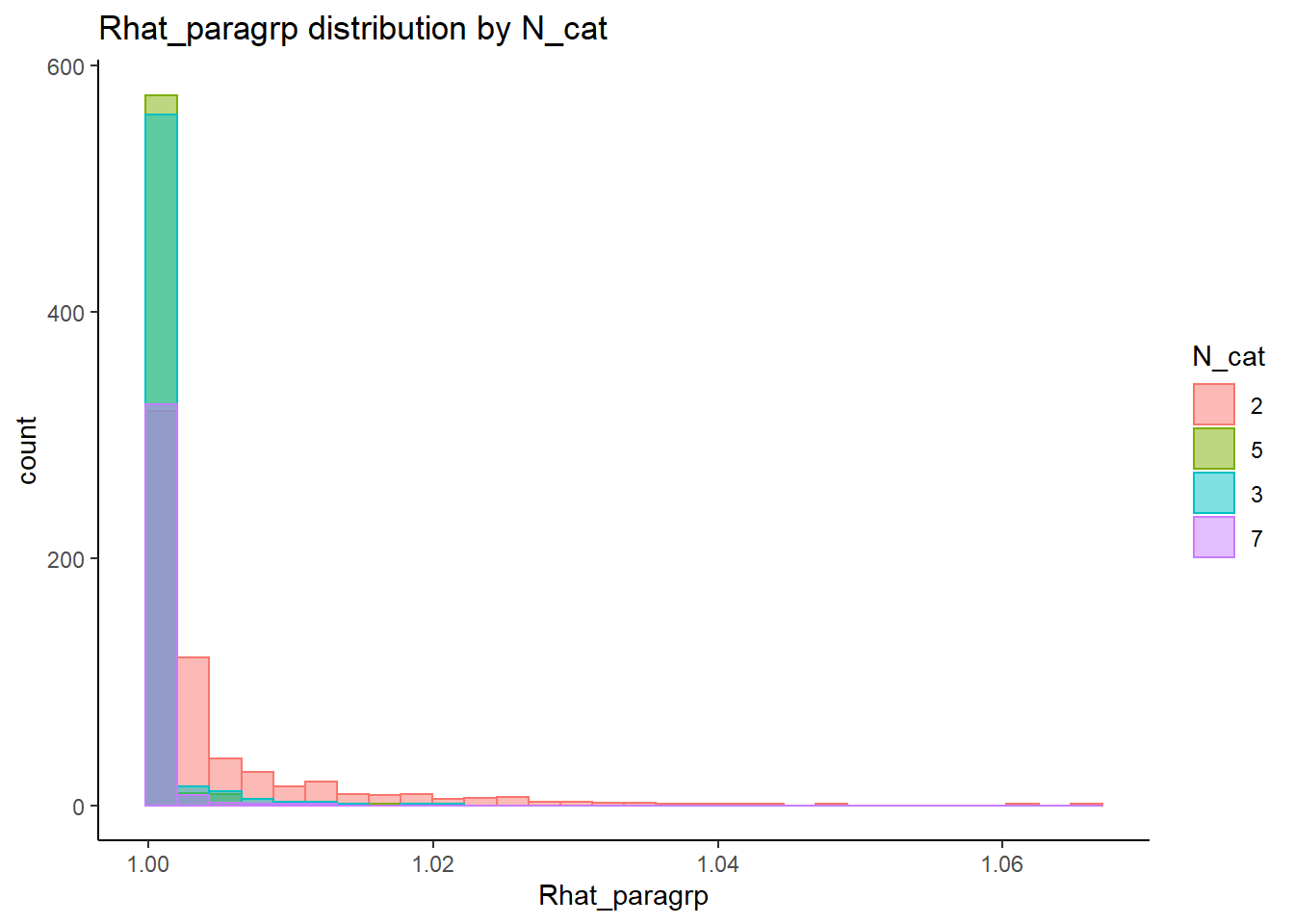
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.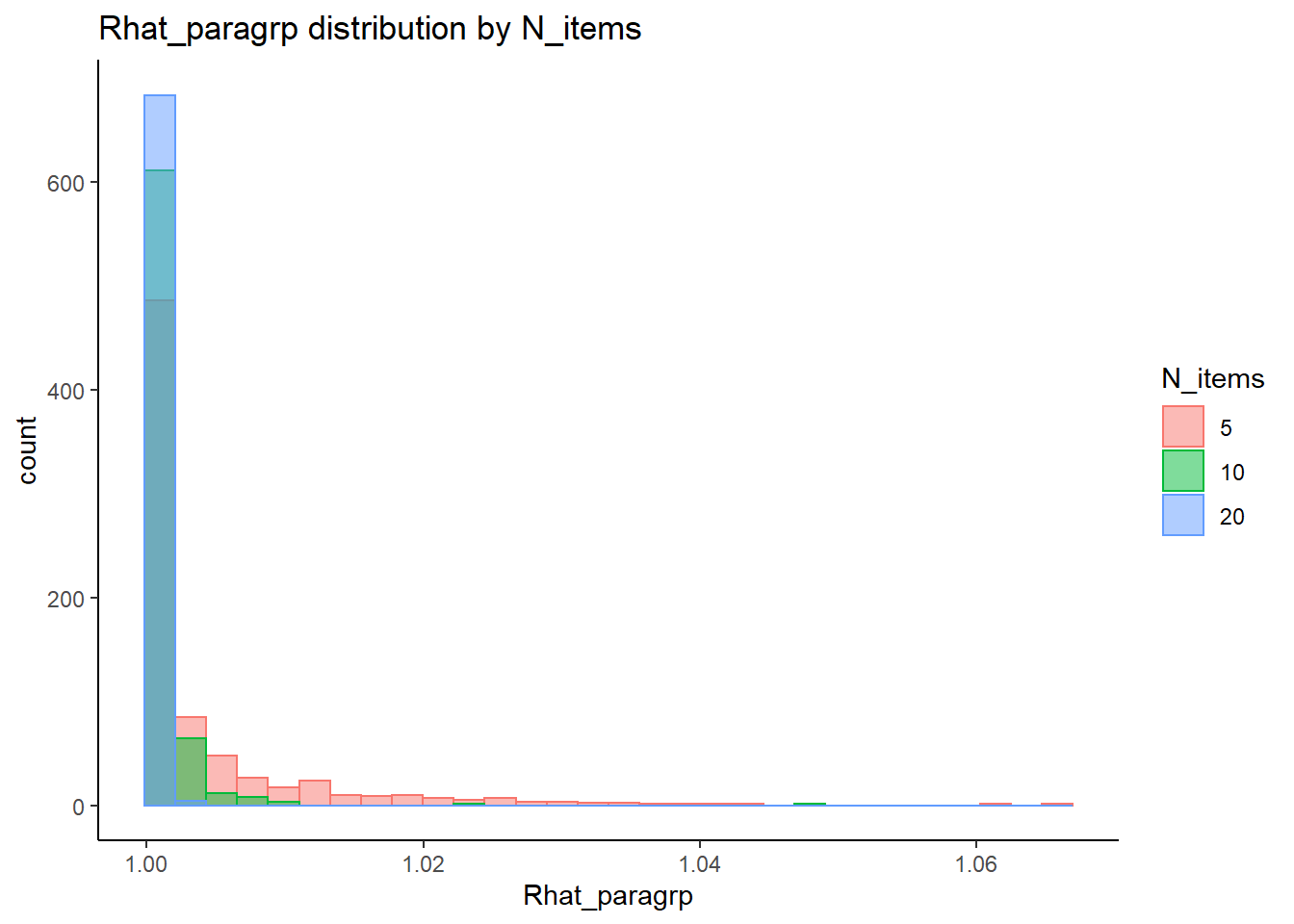
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.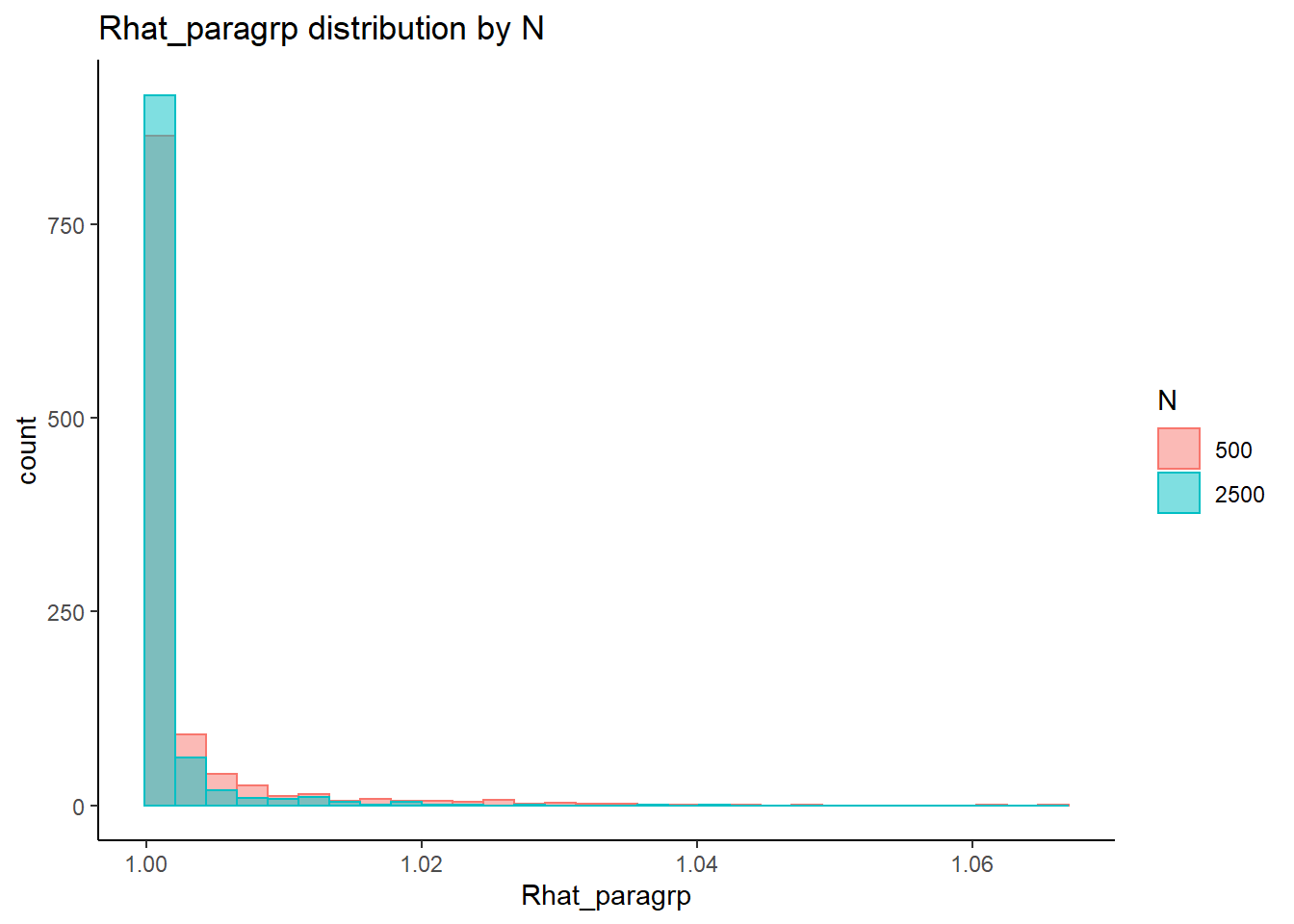
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 202 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 285 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 80 <0.0000000000000002 ***
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.00853 0.002843 231.39 < 0.0000000000000002 ***
N_items 2 0.00628 0.003140 255.63 < 0.0000000000000002 ***
N 1 0.00103 0.001026 83.53 < 0.0000000000000002 ***
N_cat:N_items 6 0.00674 0.001123 91.39 < 0.0000000000000002 ***
N_cat:N 3 0.00089 0.000296 24.07 0.0000000000000026 ***
N_items:N 2 0.00086 0.000431 35.04 0.0000000000000011 ***
N_cat:N_items:N 6 0.00034 0.000056 4.55 0.00014 ***
Residuals 2114 0.02597 0.000012
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.1677 0.2443 0.16845
N_items 0.1235 0.1924 0.12406
N 0.0200 0.0372 0.02027
N_cat:N_items 0.1316 0.2023 0.13306
N_cat:N 0.0168 0.0314 0.01752
N_items:N 0.0165 0.0309 0.01701
N_cat:N_items:N 0.0052 0.0099 0.00662saved_omega[5,] <- omega2(fit)
saved_eta[5,] <- eta_estResponse Time Factor Precision (\(\sigma_s\))
conv_para_grp <- mydata %>%
filter(parameter_group == "sigma_s") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.8, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 34.7 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 190 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 0.03 0.86
2136 fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.0167 0.00555 66.57 <0.0000000000000002 ***
N_items 2 0.0408 0.02039 244.42 <0.0000000000000002 ***
N 1 0.0000 0.00003 0.31 0.58
N_cat:N_items 6 0.0080 0.00134 16.03 <0.0000000000000002 ***
N_cat:N 3 0.0001 0.00004 0.44 0.73
N_items:N 2 0.0005 0.00023 2.82 0.06 .
N_cat:N_items:N 6 0.0003 0.00005 0.60 0.73
Residuals 2114 0.1763 0.00008
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.0676 0.0843 0.068641
N_items 0.1673 0.1855 0.168011
N -0.0002 -0.0003 0.000106
N_cat:N_items 0.0310 0.0405 0.033056
N_cat:N -0.0006 -0.0008 0.000452
N_items:N 0.0012 0.0017 0.001935
N_cat:N_items:N -0.0008 -0.0011 0.001236saved_omega[6,] <- omega2(fit)
saved_eta[6,] <- eta_estFactor Covariance (\(\sigma_{st}\))
conv_para_grp <- mydata %>%
filter(parameter_group == "sigma_st") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.7, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.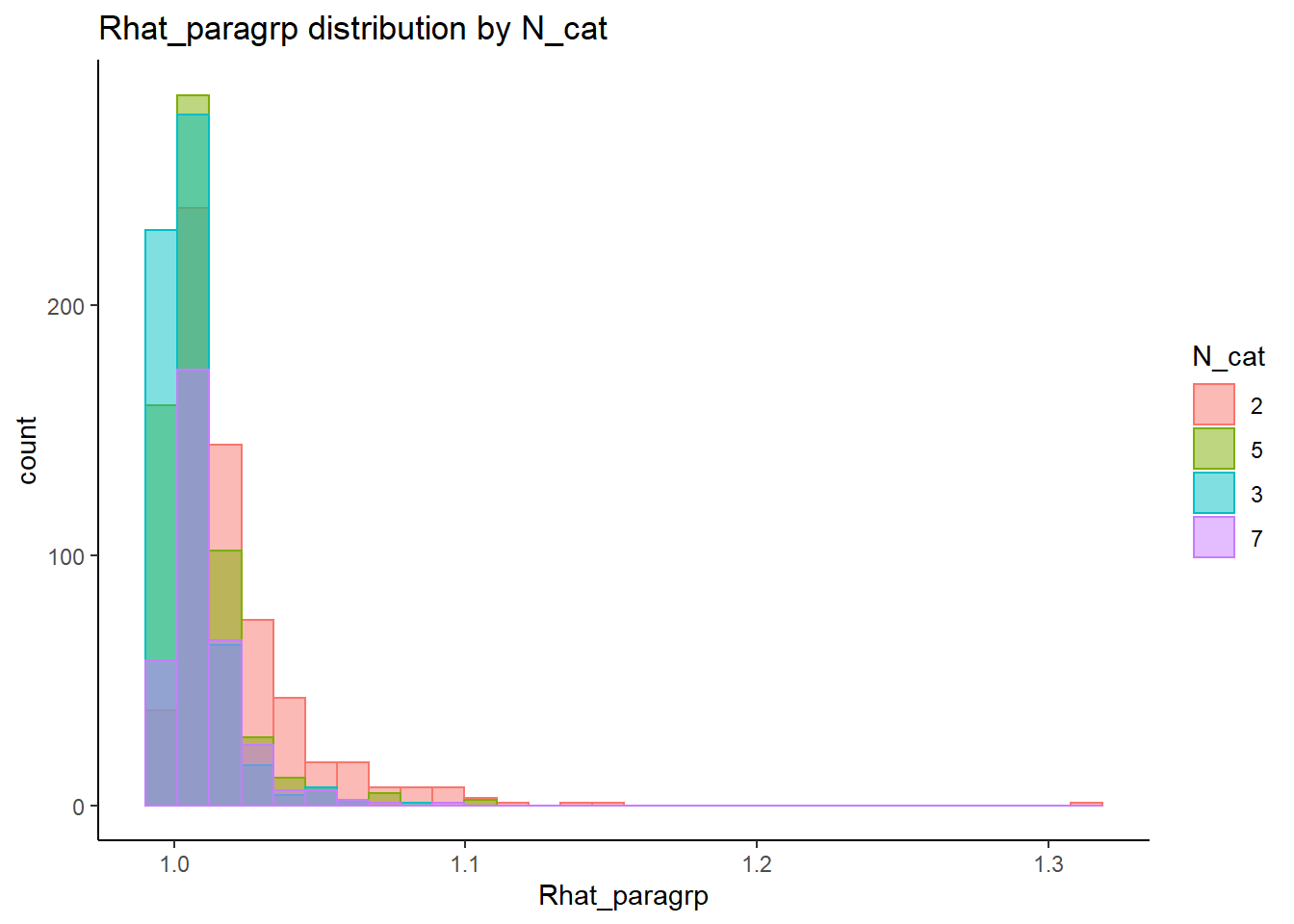
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.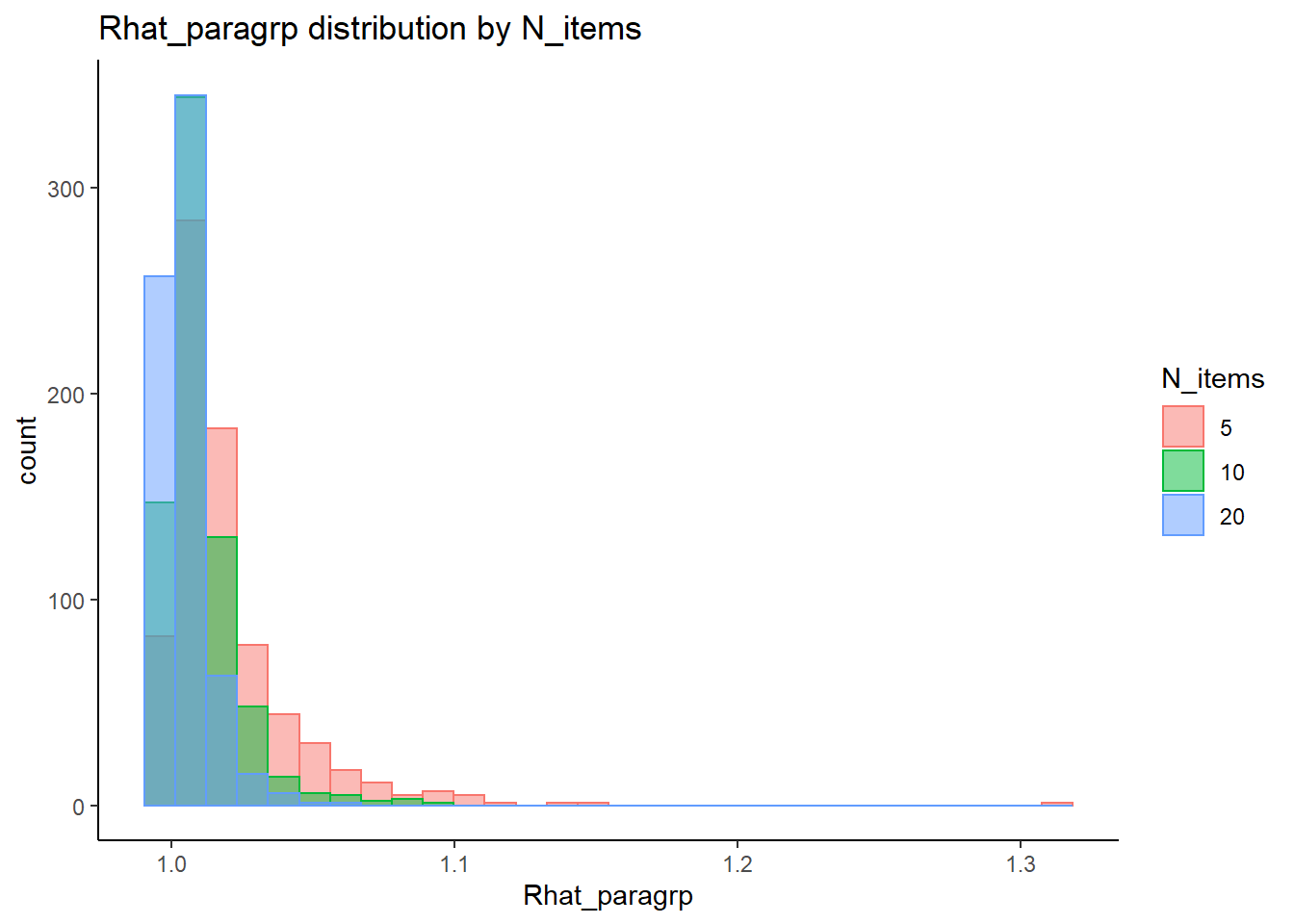
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.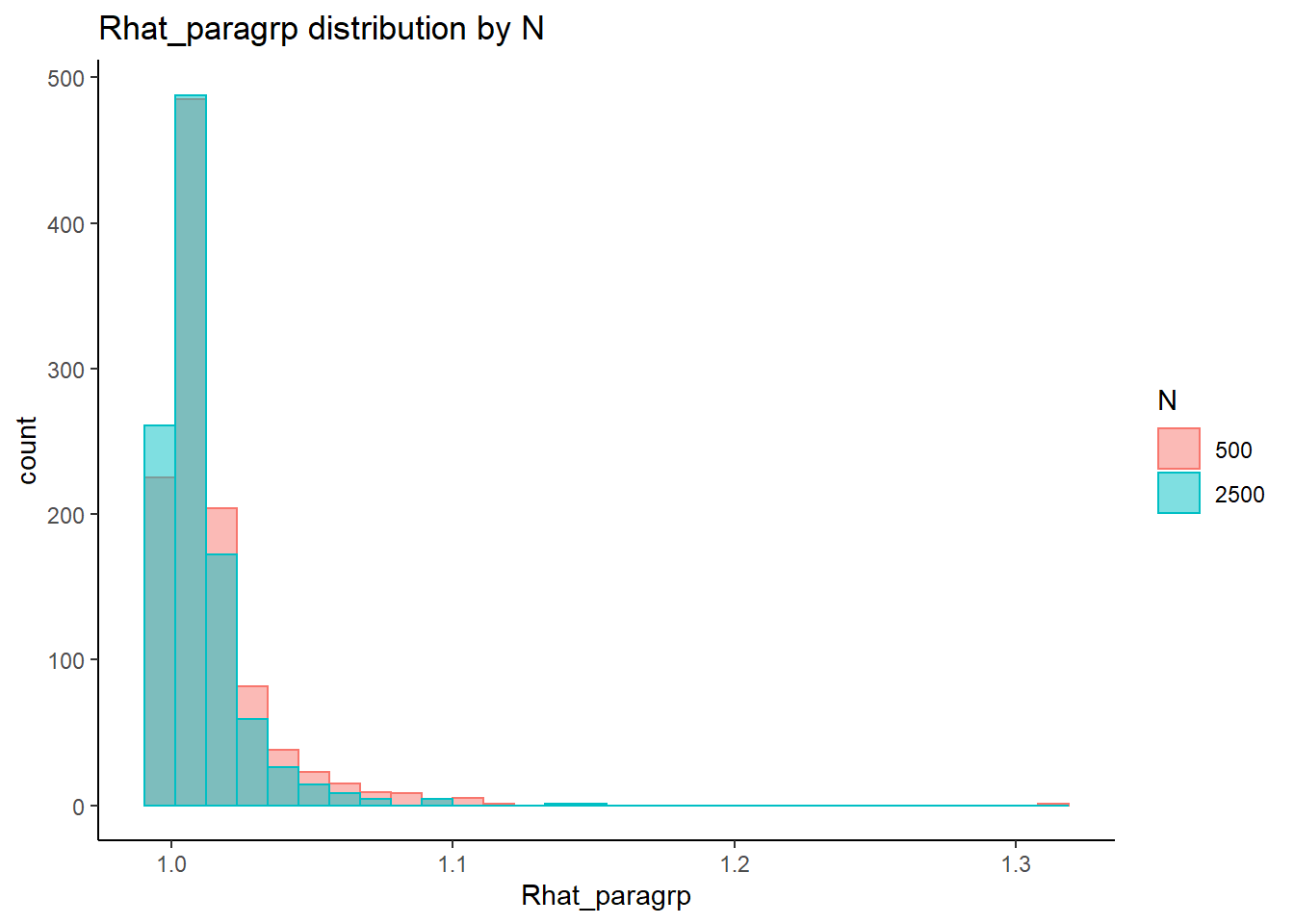
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 58.9 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 94.5 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 24.4 0.00000085 ***
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.054 0.0180 82.10 < 0.0000000000000002 ***
N_items 2 0.071 0.0353 160.59 < 0.0000000000000002 ***
N 1 0.005 0.0046 20.84 0.0000052682224 ***
N_cat:N_items 6 0.015 0.0025 11.28 0.0000000000019 ***
N_cat:N 3 0.002 0.0005 2.47 0.06 .
N_items:N 2 0.001 0.0003 1.29 0.28
N_cat:N_items:N 6 0.001 0.0001 0.49 0.81
Residuals 2114 0.464 0.0002
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.0874 0.1022 0.088504
N_items 0.1146 0.1299 0.115409
N 0.0071 0.0092 0.007490
N_cat:N_items 0.0222 0.0280 0.024326
N_cat:N 0.0016 0.0021 0.002668
N_items:N 0.0002 0.0003 0.000927
N_cat:N_items:N -0.0011 -0.0014 0.001065saved_omega[7,] <- omega2(fit)
saved_eta[7,] <- eta_estPID Relationship (\(\rho\))
conv_para_grp <- mydata %>%
filter(parameter_group == "rho") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
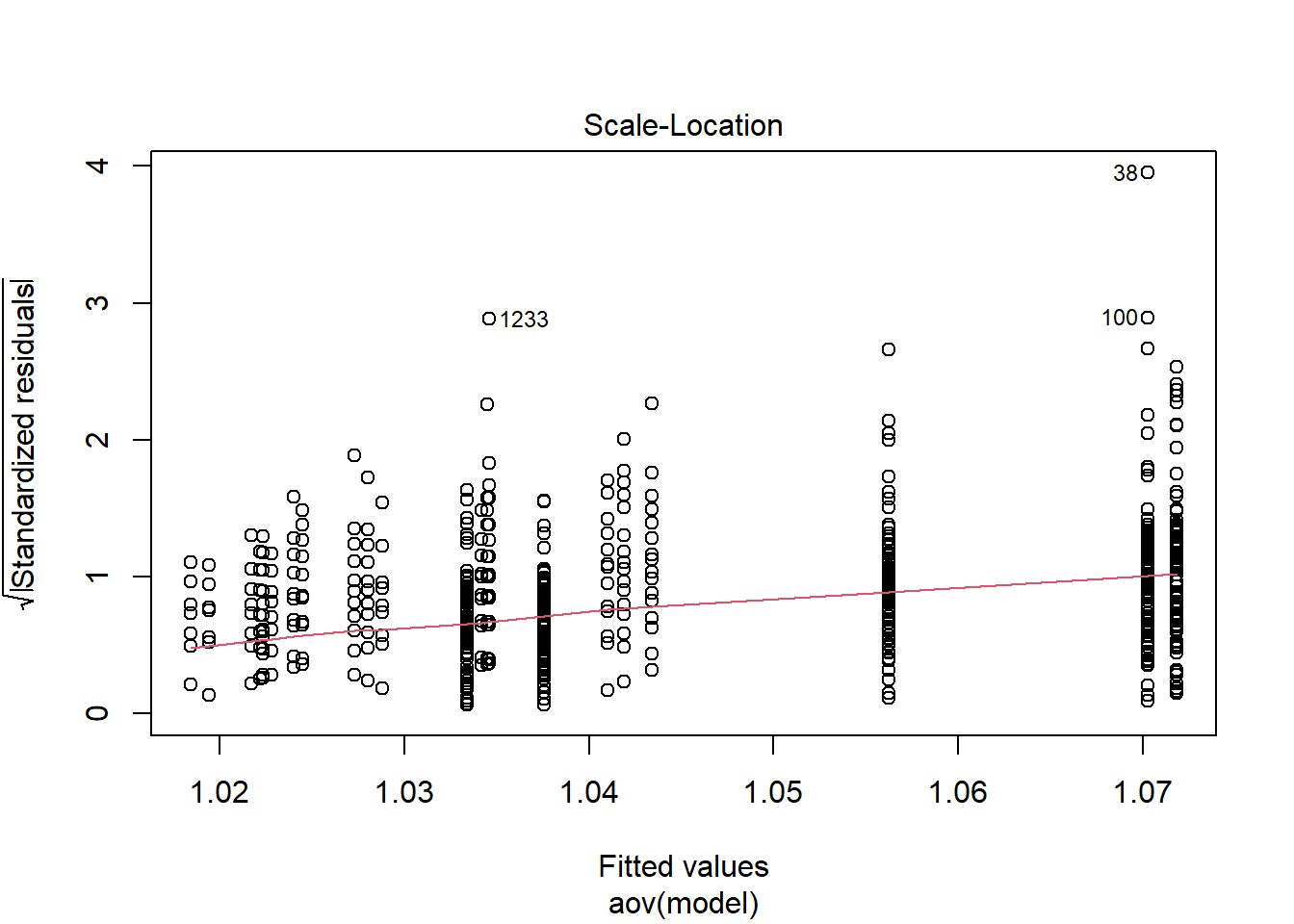
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.8, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test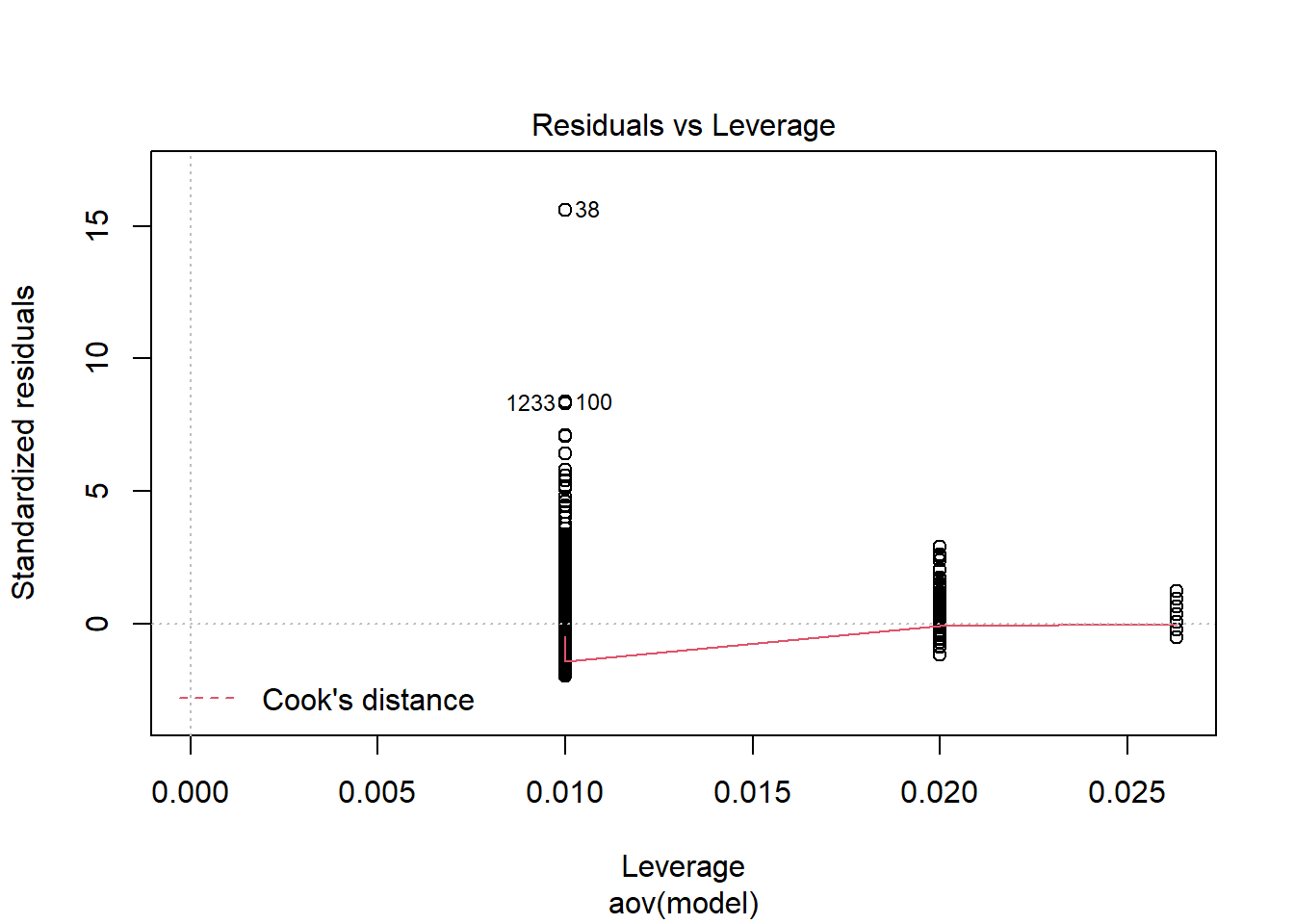
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.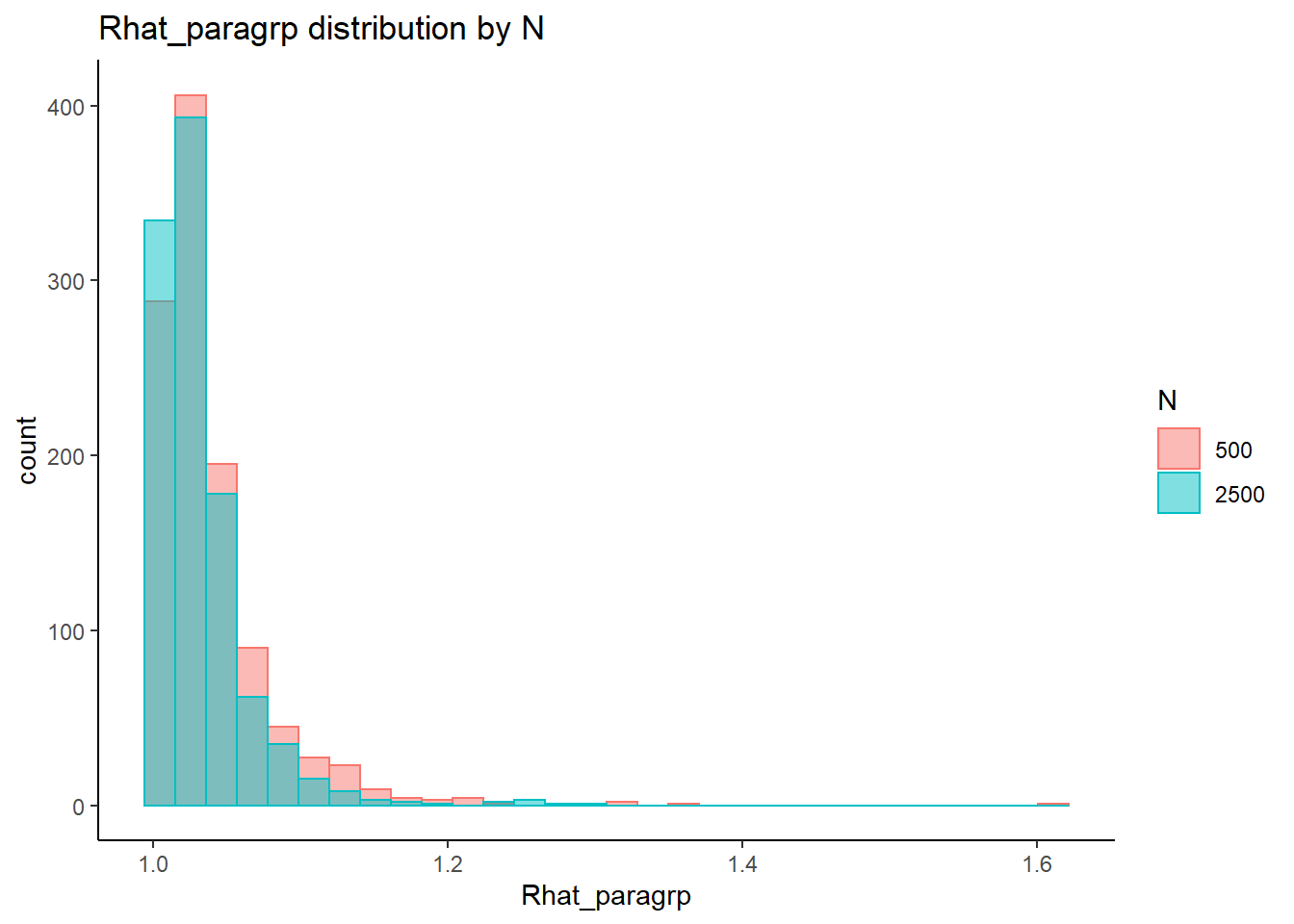
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 45.5 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 74 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 19.3 0.000012 ***
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.162 0.0540 45.25 < 0.0000000000000002 ***
N_items 2 0.185 0.0925 77.44 < 0.0000000000000002 ***
N 1 0.027 0.0273 22.88 0.000001846 ***
N_cat:N_items 6 0.053 0.0089 7.44 0.000000067 ***
N_cat:N 3 0.007 0.0024 1.99 0.114
N_items:N 2 0.008 0.0039 3.24 0.039 *
N_cat:N_items:N 6 0.010 0.0016 1.34 0.237
Residuals 2114 2.524 0.0012
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.0532 0.0585 0.05447
N_items 0.0613 0.0667 0.06213
N 0.0088 0.0101 0.00918
N_cat:N_items 0.0155 0.0178 0.01791
N_cat:N 0.0012 0.0014 0.00239
N_items:N 0.0018 0.0021 0.00260
N_cat:N_items:N 0.0008 0.0009 0.00322saved_omega[8,] <- omega2(fit)
saved_eta[8,] <- eta_estFactor Reliability (\(\omega\))
conv_para_grp <- mydata %>%
filter(parameter_group == "omega") %>%
select(all_of(keep_cols)) %>%
distinct()
## ANOVA on Rhat values
anova_assumptions_check(
dat = conv_para_grp, outcome = 'Rhat_paragrp',
factors = c('N_cat', 'N_items', 'N'),
model = as.formula('Rhat_paragrp ~ N_cat*N_items*N'))
=============================
Tests and Plots of Normality:
Shapiro-Wilks Test of Normality of Residuals:
Shapiro-Wilk normality test
data: res
W = 0.8, p-value <0.0000000000000002
K-S Test for Normality of Residuals:Warning in ks.test(aov.out$residuals, "pnorm", alternative = "two.sided"): ties
should not be present for the Kolmogorov-Smirnov test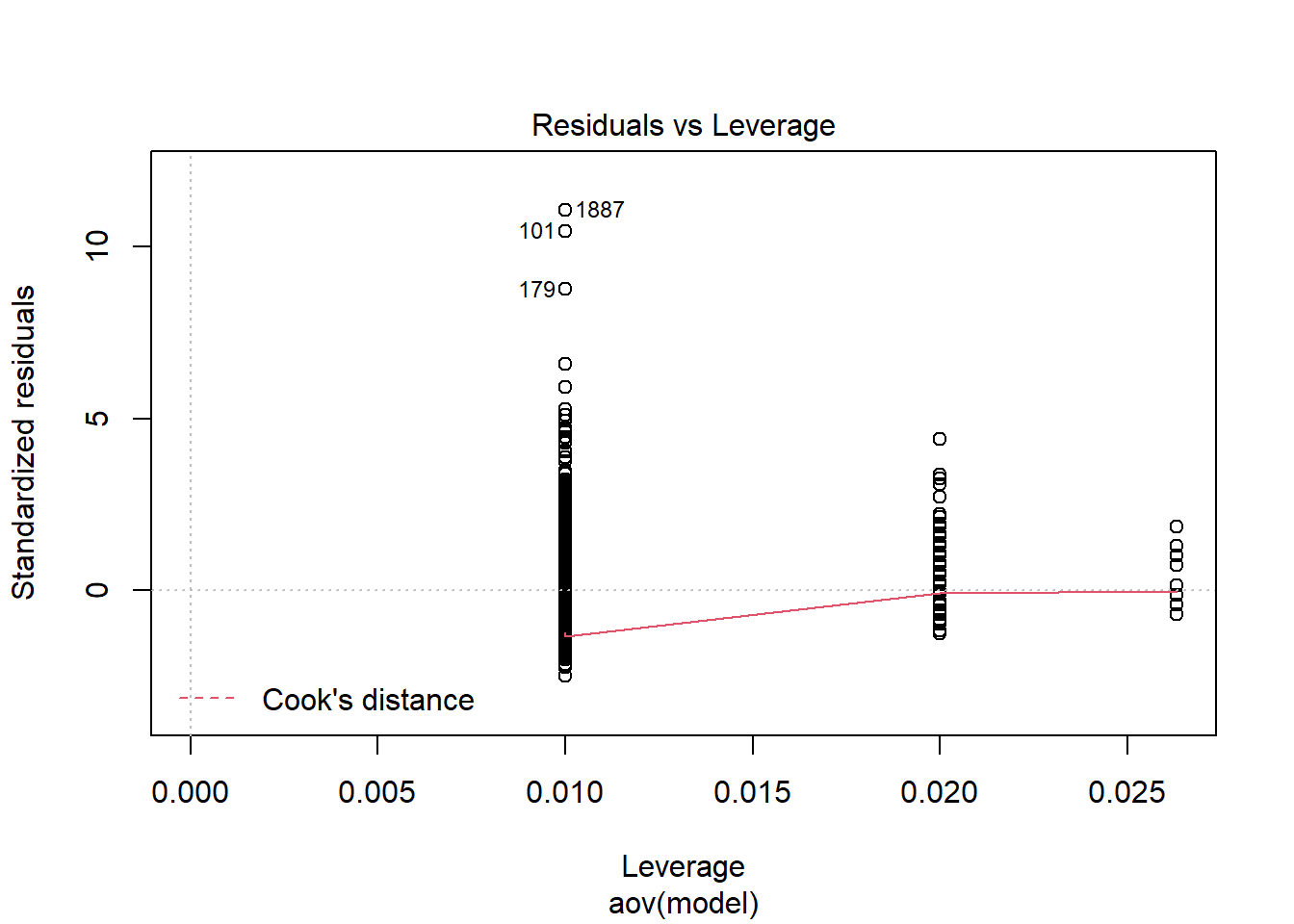
One-sample Kolmogorov-Smirnov test
data: aov.out$residuals
D = 0.5, p-value <0.0000000000000002
alternative hypothesis: two-sided`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.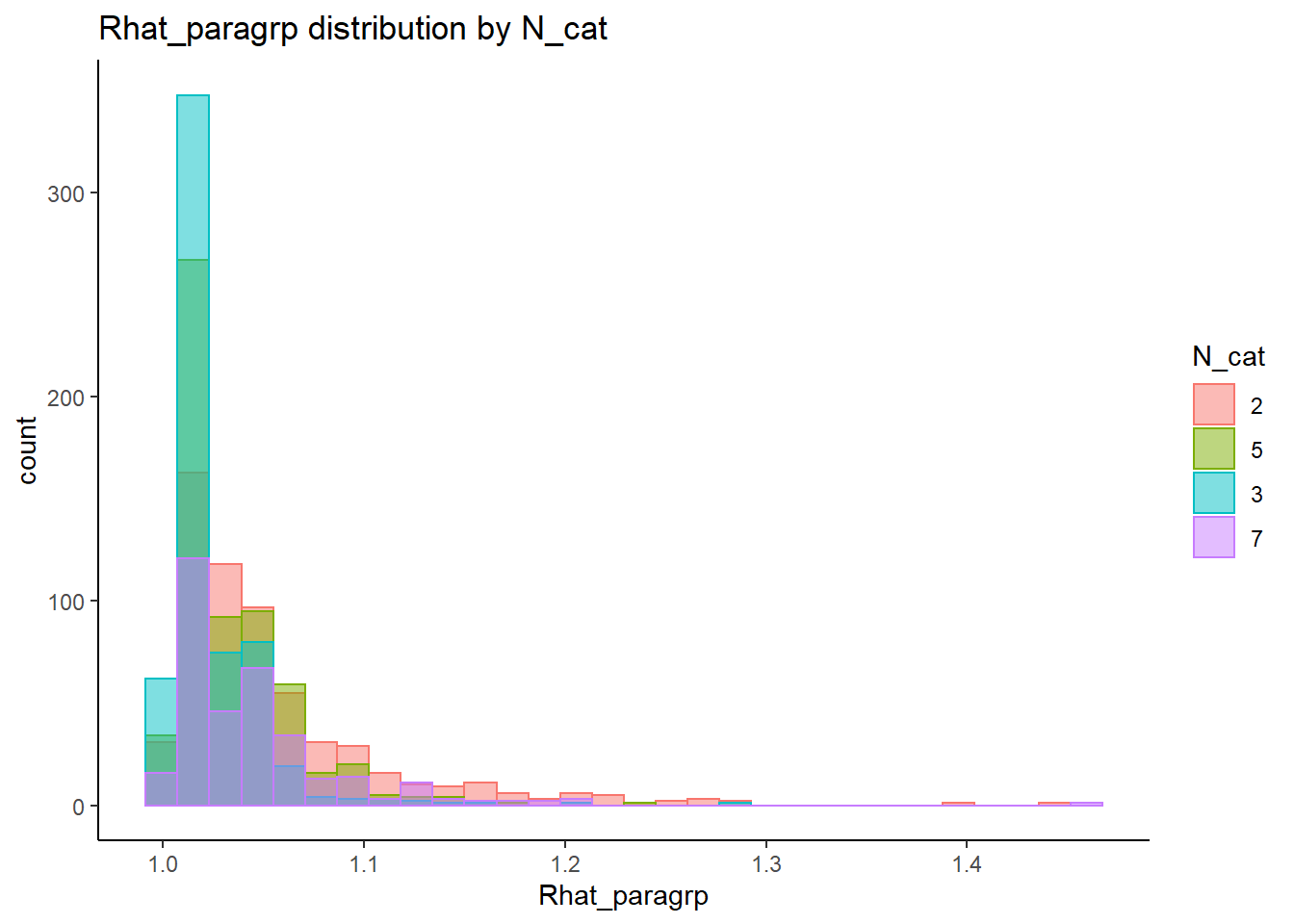
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.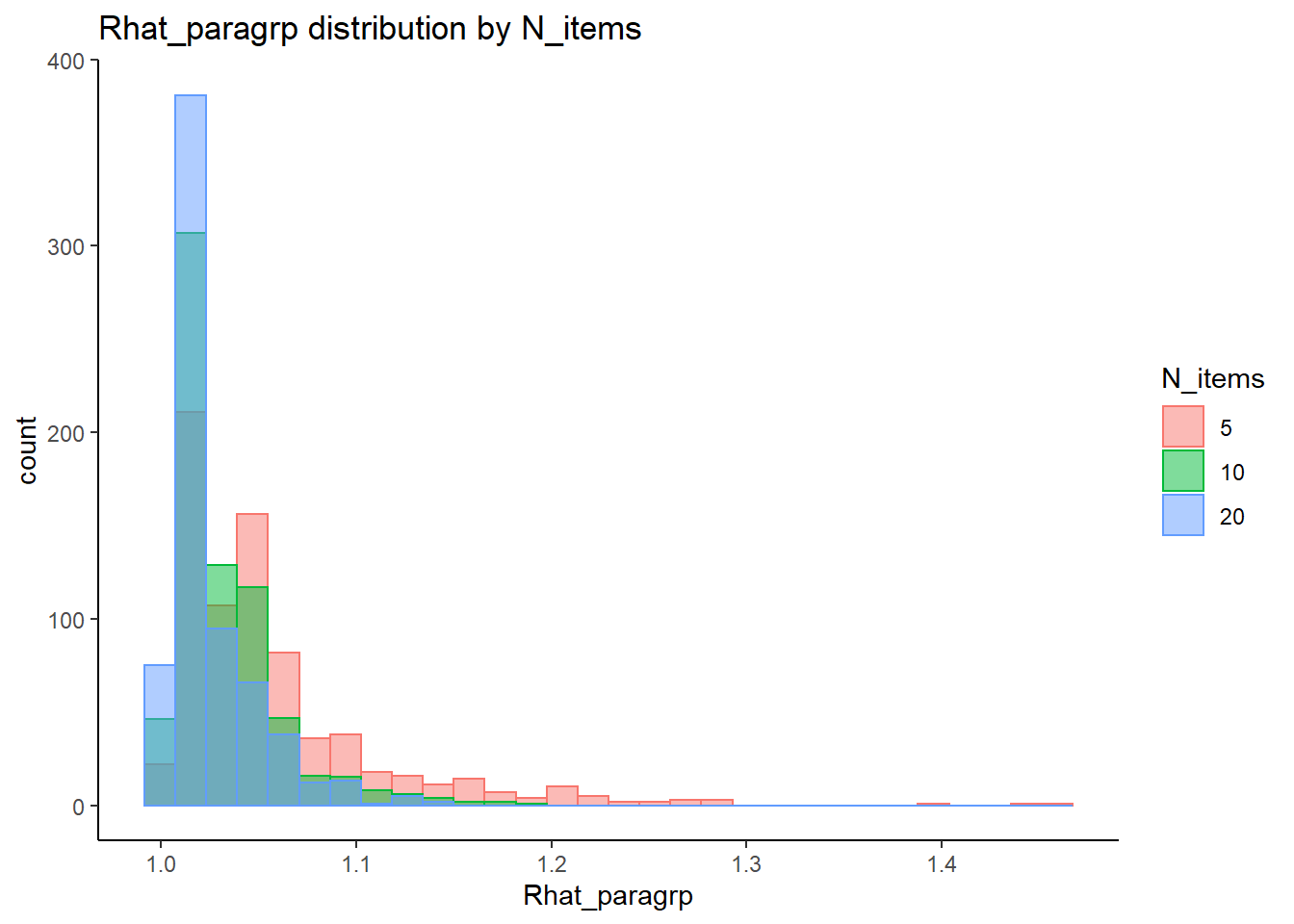
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.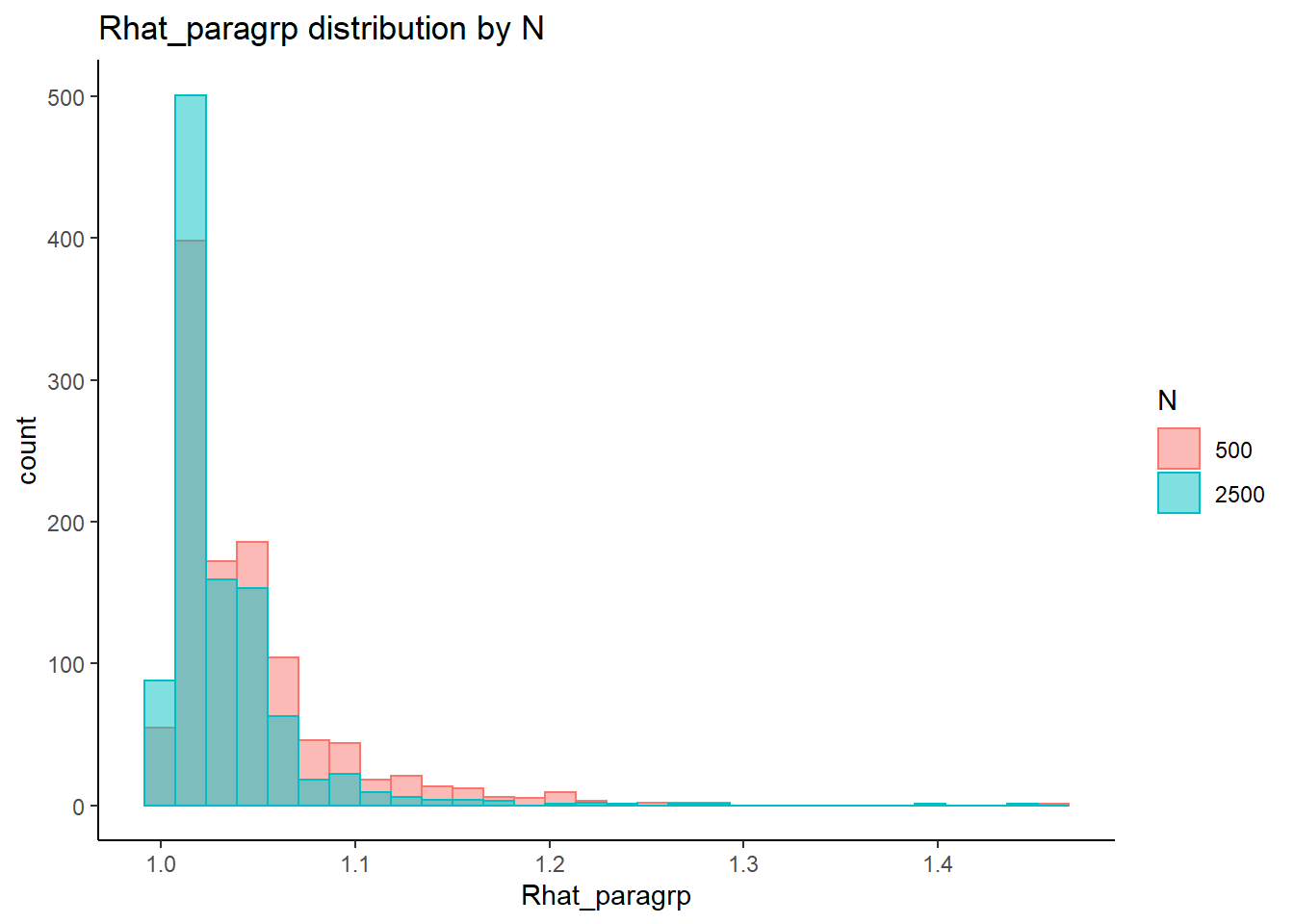
=============================
Tests of Homogeneity of Variance
Levenes Test: N_cat
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 3 67.1 <0.0000000000000002 ***
2134
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N_items
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 2 124 <0.0000000000000002 ***
2135
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Levenes Test: N
Levene's Test for Homogeneity of Variance (center = "mean")
Df F value Pr(>F)
group 1 55.7 0.00000000000012 ***
2136
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1fit <- summary(aov(Rhat_paragrp ~ N_cat*N_items*N, data=conv_para_grp))
fit Df Sum Sq Mean Sq F value Pr(>F)
N_cat 3 0.311 0.1036 80.46 < 0.0000000000000002 ***
N_items 2 0.381 0.1904 147.84 < 0.0000000000000002 ***
N 1 0.087 0.0874 67.85 0.00000000000000031 ***
N_cat:N_items 6 0.124 0.0206 16.01 < 0.0000000000000002 ***
N_cat:N 3 0.007 0.0024 1.89 0.12980
N_items:N 2 0.023 0.0113 8.78 0.00016 ***
N_cat:N_items:N 6 0.005 0.0008 0.58 0.74283
Residuals 2114 2.723 0.0013
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1eta_est <- (fit[[1]]$`Sum Sq`/sum(fit[[1]]$`Sum Sq`))[1:7]
cbind(omega2(fit),p_omega2(fit), eta_est) omega^2 partial-omega^2 eta_est
N_cat 0.0839 0.1003 0.08494
N_items 0.1033 0.1208 0.10405
N 0.0235 0.0303 0.02387
N_cat:N_items 0.0317 0.0404 0.03380
N_cat:N 0.0009 0.0012 0.00199
N_items:N 0.0055 0.0072 0.00618
N_cat:N_items:N -0.0009 -0.0012 0.00123saved_omega[9,] <- omega2(fit)
saved_eta[9,] <- eta_estManuscript Tables and Figures (latex formated)
Tables
print(
xtable(
t(saved_omega)
, caption = c("Effect of design factors on relative bias estimates: omega2")
,align = "lrrrrrrrrr", digits = 3
),
include.rownames=T,
booktabs=T
)% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Sun Feb 13 14:28:59 2022
\begin{table}[ht]
\centering
\begin{tabular}{lrrrrrrrrr}
\toprule
& lambda & tau & theta & beta\_lrt & sigma\_lrt & sigma\_s & sigma\_ts & rho & omega \\
\midrule
N\_cat & 0.189 & 0.242 & 0.186 & 0.136 & 0.168 & 0.068 & 0.087 & 0.053 & 0.084 \\
N\_items & 0.222 & 0.104 & 0.225 & 0.211 & 0.123 & 0.167 & 0.115 & 0.061 & 0.103 \\
N & 0.021 & 0.064 & 0.042 & 0.001 & 0.020 & -0.000 & 0.007 & 0.009 & 0.024 \\
N\_cat:N\_items & 0.053 & 0.026 & 0.076 & 0.053 & 0.132 & 0.031 & 0.022 & 0.015 & 0.032 \\
N\_cat:N & 0.002 & 0.003 & 0.001 & 0.000 & 0.017 & -0.001 & 0.002 & 0.001 & 0.001 \\
N\_items:N & 0.001 & 0.020 & 0.009 & 0.000 & 0.016 & 0.001 & 0.000 & 0.002 & 0.005 \\
N\_cat:N\_items:N & 0.012 & 0.000 & 0.001 & 0.001 & 0.005 & -0.001 & -0.001 & 0.001 & -0.001 \\
\bottomrule
\end{tabular}
\caption{Effect of design factors on relative bias estimates: omega2}
\end{table}print(
xtable(
t(saved_eta)
, caption = c("Effect of design factors on relative bias estimates: eta2")
,align = "lrrrrrrrrr", digits = 3
),
include.rownames=T,
booktabs=T
)% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Sun Feb 13 14:28:59 2022
\begin{table}[ht]
\centering
\begin{tabular}{lrrrrrrrrr}
\toprule
& lambda & tau & theta & beta\_lrt & sigma\_lrt & sigma\_s & sigma\_ts & rho & omega \\
\midrule
N\_cat & 0.189 & 0.243 & 0.186 & 0.137 & 0.168 & 0.069 & 0.089 & 0.054 & 0.085 \\
N\_items & 0.222 & 0.105 & 0.225 & 0.212 & 0.124 & 0.168 & 0.115 & 0.062 & 0.104 \\
N & 0.021 & 0.065 & 0.042 & 0.001 & 0.020 & 0.000 & 0.007 & 0.009 & 0.024 \\
N\_cat:N\_items & 0.054 & 0.027 & 0.077 & 0.055 & 0.133 & 0.033 & 0.024 & 0.018 & 0.034 \\
N\_cat:N & 0.002 & 0.004 & 0.002 & 0.001 & 0.018 & 0.000 & 0.003 & 0.002 & 0.002 \\
N\_items:N & 0.001 & 0.021 & 0.010 & 0.001 & 0.017 & 0.002 & 0.001 & 0.003 & 0.006 \\
N\_cat:N\_items:N & 0.014 & 0.002 & 0.002 & 0.003 & 0.007 & 0.001 & 0.001 & 0.003 & 0.001 \\
\bottomrule
\end{tabular}
\caption{Effect of design factors on relative bias estimates: eta2}
\end{table}keep_cols <- c("condition", "iter", "N", "N_items", "N_cat", "parameter_group", "Rhat_paragrp", "converge_paragrp", "converge_proportion_paragrp")
tbl_dat <- mydata %>%
select(all_of(keep_cols)) %>%
filter(
!is.na(parameter_group)
, parameter_group != "lambda (STD)"
) %>%
distinct() %>%
mutate(
prop_con = ifelse(converge_proportion_paragrp > 0.5, ">50% Converged", "<=50% Converged")
)
# get average Rhat and SD for each condition and parameter group
sum_tab <- tbl_dat %>%
group_by(N_cat, N_items, N, parameter_group) %>%
summarise(
AvgRhat = mean(Rhat_paragrp),
SDRhat = sd(Rhat_paragrp),
AvgPropConv = mean(converge_proportion_paragrp)
)`summarise()` has grouped output by 'N_cat', 'N_items', 'N'. You can override using the `.groups` argument.# table 1: 2 category data
T1 <- sum_tab %>%
filter(N_cat == 2) %>%
pivot_wider(
id_cols = c("parameter_group", "N")
, names_from = "N_items"
, values_from = all_of(c("AvgRhat", "SDRhat", "AvgPropConv"))
) %>%
arrange(.,parameter_group)
# reordering of rows to more logical for presentation
T1 <- T1[c(3:4, 15:18, 1:2, 5:8, 13:14, 11:12, 9:10) ,]
print(
xtable(
T1
, caption = c("Posterior convergence by Rhat summary: Dichotomous items")
,align = "llrrrrrrrrrr"
),
include.rownames=F,
booktabs=T
)% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Sun Feb 13 14:29:00 2022
\begin{table}[ht]
\centering
\begin{tabular}{lrrrrrrrrrr}
\toprule
parameter\_group & N & AvgRhat\_5 & AvgRhat\_10 & AvgRhat\_20 & SDRhat\_5 & SDRhat\_10 & SDRhat\_20 & AvgPropConv\_5 & AvgPropConv\_10 & AvgPropConv\_20 \\
\midrule
lambda & 500 & 1.09 & 1.06 & 1.04 & 0.05 & 0.02 & 0.01 & 0.68 & 0.85 & 0.95 \\
lambda & 2500 & 1.10 & 1.05 & 1.03 & 0.05 & 0.01 & 0.00 & 0.65 & 0.90 & 0.97 \\
tau & 500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.01 & 0.00 & 0.96 & 0.98 & 1.00 \\
tau & 2500 & 1.02 & 1.01 & 1.01 & 0.01 & 0.00 & 0.00 & 0.98 & 1.00 & 1.00 \\
theta & 500 & 1.14 & 1.08 & 1.05 & 0.07 & 0.03 & 0.01 & 0.50 & 0.76 & 0.92 \\
theta & 2500 & 1.11 & 1.05 & 1.03 & 0.05 & 0.01 & 0.01 & 0.62 & 0.89 & 0.97 \\
beta\_lrt & 500 & 1.07 & 1.04 & 1.02 & 0.04 & 0.02 & 0.01 & 0.80 & 0.93 & 0.99 \\
beta\_lrt & 2500 & 1.07 & 1.03 & 1.02 & 0.04 & 0.01 & 0.01 & 0.80 & 0.97 & 1.00 \\
sigma\_lrt & 500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 0.97 & 1.00 & 1.00 \\
sigma\_lrt & 2500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.00 & 0.00 & 0.99 & 1.00 & 1.00 \\
sigma\_s & 500 & 1.02 & 1.01 & 1.00 & 0.02 & 0.01 & 0.00 & 0.99 & 1.00 & 1.00 \\
sigma\_s & 2500 & 1.02 & 1.01 & 1.00 & 0.02 & 0.01 & 0.00 & 0.97 & 1.00 & 1.00 \\
sigma\_st & 500 & 1.04 & 1.02 & 1.01 & 0.04 & 0.02 & 0.01 & 0.95 & 1.00 & 1.00 \\
sigma\_st & 2500 & 1.03 & 1.02 & 1.01 & 0.03 & 0.01 & 0.01 & 0.98 & 1.00 & 1.00 \\
rho & 500 & 1.07 & 1.06 & 1.04 & 0.08 & 0.05 & 0.02 & 0.82 & 0.85 & 0.97 \\
rho & 2500 & 1.07 & 1.03 & 1.02 & 0.07 & 0.03 & 0.01 & 0.80 & 0.97 & 1.00 \\
omega & 500 & 1.10 & 1.05 & 1.03 & 0.07 & 0.04 & 0.03 & 0.63 & 0.88 & 0.96 \\
omega & 2500 & 1.08 & 1.03 & 1.02 & 0.08 & 0.02 & 0.02 & 0.78 & 0.97 & 1.00 \\
\bottomrule
\end{tabular}
\caption{Posterior convergence by Rhat summary: Dichotomous items}
\end{table}# table 2: 5 category data
T2 <- sum_tab %>%
filter(N_cat == 5) %>%
pivot_wider(
id_cols = c("parameter_group", "N")
, names_from = "N_items"
, values_from = all_of(c("AvgRhat", "SDRhat", "AvgPropConv"))
) %>%
arrange(.,parameter_group)
# reordering of rows to more logical for presentation
T2 <- T2[c(3:4, 15:18, 1:2, 5:8, 13:14, 11:12, 9:10) ,]
print(
xtable(
T2
, caption = c("Posterior convergence by Rhat summary: Five category items")
,align = "llrrrrrrrrrr"
),
include.rownames=F,
booktabs=T
)% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Sun Feb 13 14:29:00 2022
\begin{table}[ht]
\centering
\begin{tabular}{lrrrrrrrrrr}
\toprule
parameter\_group & N & AvgRhat\_5 & AvgRhat\_10 & AvgRhat\_20 & SDRhat\_5 & SDRhat\_10 & SDRhat\_20 & AvgPropConv\_5 & AvgPropConv\_10 & AvgPropConv\_20 \\
\midrule
lambda & 500 & 1.07 & 1.04 & 1.04 & 0.03 & 0.01 & 0.01 & 0.78 & 0.92 & 0.94 \\
lambda & 2500 & 1.04 & 1.03 & 1.03 & 0.02 & 0.01 & 0.01 & 0.91 & 0.96 & 0.97 \\
tau & 500 & 1.02 & 1.02 & 1.02 & 0.01 & 0.00 & 0.00 & 0.97 & 0.99 & 1.00 \\
tau & 2500 & 1.02 & 1.02 & 1.02 & 0.01 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
theta & 500 & 1.08 & 1.05 & 1.04 & 0.04 & 0.01 & 0.01 & 0.73 & 0.91 & 0.94 \\
theta & 2500 & 1.05 & 1.03 & 1.03 & 0.02 & 0.01 & 0.01 & 0.90 & 0.96 & 0.97 \\
beta\_lrt & 500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.02 & 0.01 & 0.93 & 0.98 & 1.00 \\
beta\_lrt & 2500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.01 & 0.01 & 0.98 & 1.00 & 1.00 \\
sigma\_lrt & 500 & 1.00 & 1.00 & 1.00 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_lrt & 2500 & 1.00 & 1.00 & 1.00 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_s & 500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_s & 2500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_st & 500 & 1.02 & 1.01 & 1.01 & 0.02 & 0.01 & 0.01 & 0.98 & 1.00 & 1.00 \\
sigma\_st & 2500 & 1.02 & 1.01 & 1.01 & 0.01 & 0.01 & 0.01 & 1.00 & 1.00 & 1.00 \\
rho & 500 & 1.04 & 1.03 & 1.03 & 0.04 & 0.03 & 0.02 & 0.91 & 0.94 & 0.98 \\
rho & 2500 & 1.03 & 1.02 & 1.02 & 0.03 & 0.02 & 0.02 & 0.97 & 1.00 & 1.00 \\
omega & 500 & 1.05 & 1.04 & 1.03 & 0.04 & 0.03 & 0.03 & 0.87 & 0.95 & 0.98 \\
omega & 2500 & 1.03 & 1.03 & 1.02 & 0.03 & 0.02 & 0.02 & 0.99 & 0.97 & 0.99 \\
\bottomrule
\end{tabular}
\caption{Posterior convergence by Rhat summary: Five category items}
\end{table}# table 3: 3 category data
T3 <- sum_tab %>%
filter(N_cat == 3) %>%
pivot_wider(
id_cols = c("parameter_group", "N")
, names_from = "N_items"
, values_from = all_of(c("AvgRhat", "SDRhat", "AvgPropConv"))
) %>%
arrange(.,parameter_group)
# reordering of rows to more logical for presentation
T3 <- T3[c(3:4, 15:18, 1:2, 5:8, 13:14, 11:12, 9:10) ,]
print(
xtable(
T3
, caption = c("Posterior convergence by Rhat summary: Three category items")
,align = "llrrrrrrrrrr"
),
include.rownames=F,
booktabs=T
)% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Sun Feb 13 14:29:00 2022
\begin{table}[ht]
\centering
\begin{tabular}{lrrrrrrrrrr}
\toprule
parameter\_group & N & AvgRhat\_5 & AvgRhat\_10 & AvgRhat\_20 & SDRhat\_5 & SDRhat\_10 & SDRhat\_20 & AvgPropConv\_5 & AvgPropConv\_10 & AvgPropConv\_20 \\
\midrule
lambda & 500 & 1.06 & 1.03 & 1.02 & 0.03 & 0.01 & 0.00 & 0.87 & 0.98 & 0.99 \\
lambda & 2500 & 1.04 & 1.02 & 1.02 & 0.01 & 0.01 & 0.00 & 0.95 & 0.99 & 1.00 \\
tau & 500 & 1.02 & 1.01 & 1.01 & 0.01 & 0.00 & 0.00 & 0.98 & 1.00 & 1.00 \\
tau & 2500 & 1.01 & 1.01 & 1.01 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
theta & 500 & 1.07 & 1.03 & 1.02 & 0.04 & 0.01 & 0.00 & 0.80 & 0.98 & 0.99 \\
theta & 2500 & 1.04 & 1.02 & 1.02 & 0.02 & 0.01 & 0.00 & 0.93 & 0.99 & 1.00 \\
beta\_lrt & 500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.01 & 0.01 & 0.96 & 1.00 & 1.00 \\
beta\_lrt & 2500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.01 & 0.01 & 0.99 & 1.00 & 1.00 \\
sigma\_lrt & 500 & 1.00 & 1.00 & 1.00 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_lrt & 2500 & 1.00 & 1.00 & 1.00 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_s & 500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_s & 2500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_st & 500 & 1.01 & 1.01 & 1.01 & 0.01 & 0.01 & 0.01 & 1.00 & 1.00 & 1.00 \\
sigma\_st & 2500 & 1.01 & 1.01 & 1.00 & 0.01 & 0.01 & 0.01 & 1.00 & 1.00 & 1.00 \\
rho & 500 & 1.03 & 1.03 & 1.02 & 0.04 & 0.02 & 0.02 & 0.97 & 0.99 & 0.99 \\
rho & 2500 & 1.03 & 1.02 & 1.02 & 0.02 & 0.02 & 0.01 & 0.98 & 1.00 & 1.00 \\
omega & 500 & 1.05 & 1.02 & 1.02 & 0.04 & 0.01 & 0.01 & 0.89 & 1.00 & 1.00 \\
omega & 2500 & 1.02 & 1.02 & 1.01 & 0.02 & 0.01 & 0.02 & 1.00 & 1.00 & 0.99 \\
\bottomrule
\end{tabular}
\caption{Posterior convergence by Rhat summary: Three category items}
\end{table}# table 4: 7 category data
T4 <- sum_tab %>%
filter(N_cat == 7) %>%
pivot_wider(
id_cols = c("parameter_group", "N")
, names_from = "N_items"
, values_from = all_of(c("AvgRhat", "SDRhat", "AvgPropConv"))
) %>%
arrange(.,parameter_group)
# reordering of rows to more logical for presentation
T4 <- T4[c(3:4, 15:18, 1:2, 5:8, 13:14, 11:12, 9:10) ,]
print(
xtable(
T4
, caption = c("Posterior convergence by Rhat summary: Seven category items")
,align = "llrrrrrrrrrr"
),
include.rownames=F,
booktabs=T
)% latex table generated in R 4.0.5 by xtable 1.8-4 package
% Sun Feb 13 14:29:00 2022
\begin{table}[ht]
\centering
\begin{tabular}{lrrrrrrrrrr}
\toprule
parameter\_group & N & AvgRhat\_5 & AvgRhat\_10 & AvgRhat\_20 & SDRhat\_5 & SDRhat\_10 & SDRhat\_20 & AvgPropConv\_5 & AvgPropConv\_10 & AvgPropConv\_20 \\
\midrule
lambda & 500 & 1.07 & 1.05 & 1.05 & 0.03 & 0.02 & 0.01 & 0.78 & 0.85 & 0.88 \\
lambda & 2500 & 1.06 & 1.05 & 1.04 & 0.02 & 0.01 & 0.01 & 0.83 & 0.90 & 0.92 \\
tau & 500 & 1.03 & 1.02 & 1.02 & 0.01 & 0.00 & 0.00 & 0.95 & 0.99 & 0.99 \\
tau & 2500 & 1.02 & 1.02 & 1.02 & 0.01 & 0.00 & 0.00 & 0.99 & 1.00 & 1.00 \\
theta & 500 & 1.08 & 1.06 & 1.05 & 0.03 & 0.02 & 0.01 & 0.75 & 0.84 & 0.86 \\
theta & 2500 & 1.06 & 1.05 & 1.04 & 0.03 & 0.01 & 0.01 & 0.82 & 0.90 & 0.92 \\
beta\_lrt & 500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.01 & 0.01 & 0.96 & 0.99 & 1.00 \\
beta\_lrt & 2500 & 1.03 & 1.02 & 1.01 & 0.02 & 0.01 & 0.01 & 0.95 & 0.99 & 1.00 \\
sigma\_lrt & 500 & 1.00 & 1.00 & 1.00 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_lrt & 2500 & 1.00 & 1.00 & 1.00 & 0.00 & 0.00 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_s & 500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_s & 2500 & 1.01 & 1.00 & 1.00 & 0.01 & 0.01 & 0.00 & 1.00 & 1.00 & 1.00 \\
sigma\_st & 500 & 1.02 & 1.01 & 1.01 & 0.02 & 0.01 & 0.01 & 1.00 & 1.00 & 1.00 \\
sigma\_st & 2500 & 1.02 & 1.01 & 1.01 & 0.01 & 0.01 & 0.01 & 1.00 & 1.00 & 1.00 \\
rho & 500 & 1.04 & 1.03 & 1.02 & 0.04 & 0.02 & 0.01 & 0.87 & 0.98 & 1.00 \\
rho & 2500 & 1.04 & 1.02 & 1.02 & 0.03 & 0.02 & 0.01 & 0.90 & 0.98 & 1.00 \\
omega & 500 & 1.07 & 1.04 & 1.03 & 0.06 & 0.03 & 0.03 & 0.81 & 0.92 & 0.98 \\
omega & 2500 & 1.04 & 1.03 & 1.02 & 0.04 & 0.02 & 0.02 & 0.86 & 0.98 & 1.00 \\
\bottomrule
\end{tabular}
\caption{Posterior convergence by Rhat summary: Seven category items}
\end{table}
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] car_3.0-10 carData_3.0-4 mvtnorm_1.1-1
[4] LaplacesDemon_16.1.4 runjags_2.2.0-2 lme4_1.1-26
[7] Matrix_1.3-2 sirt_3.9-4 R2jags_0.6-1
[10] rjags_4-12 eRm_1.0-2 diffIRT_1.5
[13] statmod_1.4.35 xtable_1.8-4 kableExtra_1.3.4
[16] lavaan_0.6-7 polycor_0.7-10 bayesplot_1.8.0
[19] ggmcmc_1.5.1.1 coda_0.19-4 data.table_1.14.0
[22] patchwork_1.1.1 forcats_0.5.1 stringr_1.4.0
[25] dplyr_1.0.5 purrr_0.3.4 readr_1.4.0
[28] tidyr_1.1.3 tibble_3.1.0 ggplot2_3.3.5
[31] tidyverse_1.3.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] minqa_1.2.4 TAM_3.5-19 colorspace_2.0-0 rio_0.5.26
[5] ellipsis_0.3.1 ggridges_0.5.3 rprojroot_2.0.2 fs_1.5.0
[9] rstudioapi_0.13 farver_2.1.0 fansi_0.4.2 lubridate_1.7.10
[13] xml2_1.3.2 splines_4.0.5 mnormt_2.0.2 knitr_1.31
[17] jsonlite_1.7.2 nloptr_1.2.2.2 broom_0.7.5 dbplyr_2.1.0
[21] compiler_4.0.5 httr_1.4.2 backports_1.2.1 assertthat_0.2.1
[25] cli_2.3.1 later_1.1.0.1 htmltools_0.5.1.1 tools_4.0.5
[29] gtable_0.3.0 glue_1.4.2 Rcpp_1.0.7 cellranger_1.1.0
[33] jquerylib_0.1.3 vctrs_0.3.6 svglite_2.0.0 nlme_3.1-152
[37] psych_2.0.12 xfun_0.21 ps_1.6.0 openxlsx_4.2.3
[41] rvest_1.0.0 lifecycle_1.0.0 MASS_7.3-53.1 scales_1.1.1
[45] hms_1.0.0 promises_1.2.0.1 parallel_4.0.5 RColorBrewer_1.1-2
[49] curl_4.3 yaml_2.2.1 sass_0.3.1 reshape_0.8.8
[53] stringi_1.5.3 highr_0.8 zip_2.1.1 boot_1.3-27
[57] rlang_0.4.10 pkgconfig_2.0.3 systemfonts_1.0.1 evaluate_0.14
[61] lattice_0.20-41 labeling_0.4.2 tidyselect_1.1.0 GGally_2.1.1
[65] plyr_1.8.6 magrittr_2.0.1 R6_2.5.0 generics_0.1.0
[69] DBI_1.1.1 foreign_0.8-81 pillar_1.5.1 haven_2.3.1
[73] withr_2.4.1 abind_1.4-5 modelr_0.1.8 crayon_1.4.1
[77] utf8_1.1.4 tmvnsim_1.0-2 rmarkdown_2.7 grid_4.0.5
[81] readxl_1.3.1 CDM_7.5-15 pbivnorm_0.6.0 git2r_0.28.0
[85] reprex_1.0.0 digest_0.6.27 webshot_0.5.2 httpuv_1.5.5
[89] stats4_4.0.5 munsell_0.5.0 viridisLite_0.3.0 bslib_0.2.4
[93] R2WinBUGS_2.1-21