POOLS Development: Exploratory Factor Analysis
Shan Jiang, Laura Shero, R. Noah Padgett, & Todd Kettler
2020-11-19
Last updated: 2021-02-18
Checks: 7 0
Knit directory: pools-projects/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201007) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5934257. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: manuscript/pools-development-paper-2021-02-18-blind.docx
Untracked: manuscript/pools-development-paper-2021-02-18-title-page.docx
Unstaged changes:
Modified: .Rprofile
Deleted: .gitattributes
Modified: .gitignore
Modified: manuscript/pools-development-paper-2021-02-18.docx
Modified: manuscript/~$ols-development-paper-2021-02-18.docx
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/development-study-EFA.Rmd) and HTML (docs/development-study-EFA.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5934257 | noah-padgett | 2021-02-18 | major revision to structure |
| html | 5934257 | noah-padgett | 2021-02-18 | major revision to structure |
Prior Expectations
Prior to conducting the EFA, we expect that the four factors will emerge from a four-factor solution.
Data
source("code/load_packages.R")
mydata <- read.table("data/data-2020-11-16/pools_data_split1_2020_11_16.txt", sep="\t", header=T)
dat <- mydata[,7:63]
cov.dat <- cov(dat)
# setup output object
out.dat <- data.frame(matrix(0, nrow=57, ncol=10))
rownames(out.dat) <- colnames(dat)
colnames(out.dat) <- c("Diff", "SD", "ITCc", "ITCT", "f1", "f2", "f3", "f4", "h2", "u2")Item Analysis
For the basic item analysis, we conducted these by the a priori item groups to get a sense of how the initially proposed items fit together.
All Items
fit <- psychometric::item.exam(dat)
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q4_1 | 0.866 | 0.638 | 0.622 | 2.460 | 0.552 | 0.538 |
| Q4_2 | 0.819 | 0.681 | 0.667 | 2.305 | 0.557 | 0.546 |
| Q4_3 | 0.834 | 0.645 | 0.631 | 2.537 | 0.538 | 0.525 |
| Q4_4 | 0.816 | 0.705 | 0.692 | 2.640 | 0.574 | 0.564 |
| Q4_5 | 0.880 | 0.676 | 0.661 | 2.320 | 0.594 | 0.581 |
| Q4_6 | 0.981 | 0.659 | 0.642 | 2.872 | 0.645 | 0.629 |
| Q4_7 | 0.938 | 0.546 | 0.526 | 3.582 | 0.512 | 0.493 |
| Q4_8 | 0.927 | 0.655 | 0.639 | 2.287 | 0.606 | 0.591 |
| Q4_9 | 1.007 | 0.614 | 0.595 | 2.463 | 0.617 | 0.598 |
| Q4_10 | 0.789 | 0.663 | 0.650 | 2.582 | 0.523 | 0.512 |
| Q4_11 | 0.955 | 0.729 | 0.715 | 2.500 | 0.694 | 0.681 |
| Q4_12 | 1.051 | 0.653 | 0.635 | 2.195 | 0.685 | 0.666 |
| Q4_13 | 0.876 | 0.584 | 0.566 | 1.595 | 0.511 | 0.495 |
| Q4_14 | 0.895 | 0.697 | 0.683 | 2.659 | 0.622 | 0.610 |
| Q4_15 | 0.917 | 0.722 | 0.708 | 2.375 | 0.661 | 0.648 |
| Q4_16 | 0.941 | 0.661 | 0.645 | 2.457 | 0.621 | 0.606 |
| Q4_17 | 0.964 | 0.631 | 0.614 | 2.198 | 0.608 | 0.591 |
| Q4_18 | 0.823 | 0.737 | 0.726 | 2.360 | 0.606 | 0.597 |
| Q4_19 | 0.970 | 0.733 | 0.719 | 2.543 | 0.709 | 0.696 |
| Q5_1 | 0.951 | 0.652 | 0.635 | 2.637 | 0.619 | 0.603 |
| Q5_2 | 1.035 | 0.643 | 0.624 | 2.954 | 0.664 | 0.645 |
| Q5_3 | 1.046 | 0.649 | 0.631 | 2.662 | 0.678 | 0.659 |
| Q5_4 | 1.106 | 0.566 | 0.543 | 3.500 | 0.625 | 0.600 |
| Q5_5 | 1.034 | 0.495 | 0.471 | 3.530 | 0.511 | 0.486 |
| Q5_6 | 0.906 | 0.686 | 0.671 | 2.927 | 0.620 | 0.607 |
| Q5_7 | 0.942 | 0.636 | 0.619 | 2.704 | 0.598 | 0.582 |
| Q5_8 | 1.055 | 0.667 | 0.649 | 2.905 | 0.703 | 0.684 |
| Q5_9 | 0.995 | 0.728 | 0.714 | 2.610 | 0.724 | 0.709 |
| Q5_10 | 0.837 | 0.610 | 0.594 | 2.622 | 0.510 | 0.496 |
| Q5_11 | 0.951 | 0.674 | 0.658 | 2.707 | 0.640 | 0.625 |
| Q5_12 | 0.952 | 0.660 | 0.644 | 2.881 | 0.627 | 0.612 |
| Q6_1 | 0.844 | 0.526 | 0.508 | 1.701 | 0.443 | 0.428 |
| Q6_2 | 0.912 | 0.581 | 0.563 | 2.079 | 0.529 | 0.512 |
| Q6_3 | 0.924 | 0.561 | 0.542 | 1.930 | 0.518 | 0.500 |
| Q6_4 | 0.927 | 0.510 | 0.490 | 2.076 | 0.473 | 0.453 |
| Q6_5 | 1.033 | 0.495 | 0.472 | 2.351 | 0.511 | 0.486 |
| Q6_6 | 0.820 | 0.641 | 0.626 | 1.832 | 0.525 | 0.513 |
| Q6_7 | 0.875 | 0.680 | 0.666 | 2.162 | 0.594 | 0.582 |
| Q6_8 | 0.903 | 0.630 | 0.613 | 2.113 | 0.568 | 0.553 |
| Q6_9 | 1.222 | 0.532 | 0.505 | 2.768 | 0.649 | 0.616 |
| Q6_10 | 0.913 | 0.593 | 0.575 | 2.887 | 0.541 | 0.524 |
| Q6_11 | 0.970 | 0.714 | 0.699 | 2.814 | 0.691 | 0.677 |
| Q7_1 | 0.901 | 0.758 | 0.746 | 2.463 | 0.682 | 0.671 |
| Q7_2 | 0.857 | 0.693 | 0.679 | 2.692 | 0.592 | 0.581 |
| Q7_3 | 1.012 | 0.575 | 0.555 | 3.558 | 0.581 | 0.561 |
| Q7_4 | 0.980 | 0.628 | 0.610 | 2.841 | 0.614 | 0.597 |
| Q7_5 | 0.948 | 0.719 | 0.705 | 2.875 | 0.681 | 0.668 |
| Q7_6 | 0.920 | 0.679 | 0.664 | 2.933 | 0.624 | 0.610 |
| Q7_7 | 1.014 | 0.598 | 0.579 | 3.652 | 0.606 | 0.586 |
| Q7_8 | 0.877 | 0.634 | 0.618 | 2.814 | 0.555 | 0.542 |
| Q7_9 | 0.993 | 0.710 | 0.695 | 2.567 | 0.704 | 0.689 |
| Q7_10 | 0.945 | 0.715 | 0.701 | 2.280 | 0.675 | 0.661 |
| Q7_11 | 0.840 | 0.671 | 0.657 | 2.744 | 0.562 | 0.550 |
| Q7_12 | 1.023 | 0.556 | 0.535 | 3.390 | 0.568 | 0.546 |
| Q7_13 | 0.989 | 0.300 | 0.273 | 3.579 | 0.296 | 0.269 |
| Q7_14 | 0.986 | 0.454 | 0.430 | 3.610 | 0.447 | 0.424 |
| Q7_15 | 0.905 | 0.663 | 0.648 | 2.509 | 0.600 | 0.586 |
out.dat[, c(1,2, 4)] <- fit[, c(4, 1, 3)]Initial Effective Learning Items
Item Analysis
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q4_",1:19)])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q4_1 | 0.866 | 0.750 | 0.717 | 2.460 | 0.649 | 0.621 |
| Q4_2 | 0.819 | 0.789 | 0.763 | 2.305 | 0.646 | 0.624 |
| Q4_3 | 0.834 | 0.744 | 0.712 | 2.537 | 0.620 | 0.593 |
| Q4_4 | 0.816 | 0.772 | 0.743 | 2.640 | 0.629 | 0.605 |
| Q4_5 | 0.880 | 0.731 | 0.696 | 2.320 | 0.643 | 0.612 |
| Q4_6 | 0.981 | 0.650 | 0.601 | 2.872 | 0.637 | 0.589 |
| Q4_7 | 0.938 | 0.515 | 0.457 | 3.582 | 0.483 | 0.428 |
| Q4_8 | 0.927 | 0.777 | 0.745 | 2.287 | 0.719 | 0.690 |
| Q4_9 | 1.007 | 0.701 | 0.657 | 2.463 | 0.705 | 0.660 |
| Q4_10 | 0.789 | 0.730 | 0.698 | 2.582 | 0.575 | 0.550 |
| Q4_11 | 0.955 | 0.792 | 0.761 | 2.500 | 0.755 | 0.725 |
| Q4_12 | 1.051 | 0.696 | 0.649 | 2.195 | 0.730 | 0.681 |
| Q4_13 | 0.876 | 0.650 | 0.607 | 1.595 | 0.569 | 0.531 |
| Q4_14 | 0.895 | 0.716 | 0.679 | 2.659 | 0.640 | 0.607 |
| Q4_15 | 0.917 | 0.789 | 0.759 | 2.375 | 0.722 | 0.695 |
| Q4_16 | 0.941 | 0.726 | 0.688 | 2.457 | 0.682 | 0.646 |
| Q4_17 | 0.964 | 0.720 | 0.680 | 2.198 | 0.694 | 0.655 |
| Q4_18 | 0.823 | 0.843 | 0.822 | 2.360 | 0.693 | 0.676 |
| Q4_19 | 0.970 | 0.793 | 0.762 | 2.543 | 0.768 | 0.738 |
out.dat[1:19, 3] <- fit[, 3]Preliminary Reliability
# preliminary reliability
psych::alpha(dat[, paste0("Q4_",1:19)])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q4_", 1:19)])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.95 0.95 0.96 0.51 20 0.004 2.5 0.66 0.51
lower alpha upper 95% confidence boundaries
0.94 0.95 0.96
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q4_1 0.95 0.95 0.96 0.51 18 0.0042 0.0103 0.51
Q4_2 0.95 0.95 0.96 0.50 18 0.0043 0.0103 0.51
Q4_3 0.95 0.95 0.96 0.51 18 0.0042 0.0101 0.51
Q4_4 0.95 0.95 0.96 0.50 18 0.0042 0.0105 0.51
Q4_5 0.95 0.95 0.96 0.51 19 0.0042 0.0109 0.51
Q4_6 0.95 0.95 0.96 0.52 19 0.0041 0.0105 0.53
Q4_7 0.95 0.95 0.96 0.53 20 0.0039 0.0069 0.53
Q4_8 0.95 0.95 0.96 0.50 18 0.0043 0.0106 0.51
Q4_9 0.95 0.95 0.96 0.51 19 0.0041 0.0104 0.52
Q4_10 0.95 0.95 0.96 0.51 19 0.0042 0.0108 0.51
Q4_11 0.95 0.95 0.96 0.50 18 0.0043 0.0108 0.51
Q4_12 0.95 0.95 0.96 0.51 19 0.0041 0.0105 0.52
Q4_13 0.95 0.95 0.96 0.52 19 0.0041 0.0094 0.52
Q4_14 0.95 0.95 0.96 0.51 19 0.0042 0.0109 0.52
Q4_15 0.95 0.95 0.96 0.50 18 0.0043 0.0103 0.51
Q4_16 0.95 0.95 0.96 0.51 19 0.0042 0.0107 0.52
Q4_17 0.95 0.95 0.96 0.51 19 0.0042 0.0108 0.52
Q4_18 0.95 0.95 0.96 0.50 18 0.0043 0.0096 0.50
Q4_19 0.95 0.95 0.96 0.50 18 0.0043 0.0105 0.51
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q4_1 328 0.75 0.76 0.74 0.72 2.5 0.87
Q4_2 328 0.79 0.80 0.79 0.76 2.3 0.82
Q4_3 328 0.74 0.75 0.74 0.71 2.5 0.83
Q4_4 328 0.77 0.78 0.77 0.74 2.6 0.82
Q4_5 328 0.73 0.73 0.72 0.70 2.3 0.88
Q4_6 328 0.65 0.65 0.62 0.60 2.9 0.98
Q4_7 328 0.52 0.51 0.48 0.46 3.6 0.94
Q4_8 328 0.78 0.78 0.76 0.75 2.3 0.93
Q4_9 328 0.70 0.70 0.68 0.66 2.5 1.01
Q4_10 328 0.73 0.74 0.72 0.70 2.6 0.79
Q4_11 328 0.79 0.79 0.78 0.76 2.5 0.95
Q4_12 328 0.70 0.69 0.67 0.65 2.2 1.05
Q4_13 328 0.65 0.65 0.63 0.61 1.6 0.88
Q4_14 328 0.72 0.72 0.70 0.68 2.7 0.89
Q4_15 328 0.79 0.79 0.78 0.76 2.4 0.92
Q4_16 328 0.73 0.72 0.71 0.69 2.5 0.94
Q4_17 328 0.72 0.72 0.70 0.68 2.2 0.96
Q4_18 328 0.84 0.85 0.84 0.82 2.4 0.82
Q4_19 328 0.79 0.79 0.78 0.76 2.5 0.97
Non missing response frequency for each item
1 2 3 4 5 miss
Q4_1 0.13 0.38 0.39 0.09 0.01 0
Q4_2 0.15 0.47 0.32 0.05 0.01 0
Q4_3 0.10 0.37 0.44 0.08 0.02 0
Q4_4 0.09 0.28 0.54 0.08 0.02 0
Q4_5 0.16 0.47 0.28 0.08 0.01 0
Q4_6 0.09 0.23 0.44 0.20 0.05 0
Q4_7 0.04 0.09 0.24 0.52 0.11 0
Q4_8 0.20 0.43 0.28 0.07 0.02 0
Q4_9 0.15 0.42 0.27 0.12 0.04 0
Q4_10 0.09 0.30 0.54 0.04 0.02 0
Q4_11 0.16 0.34 0.38 0.10 0.02 0
Q4_12 0.28 0.40 0.18 0.11 0.03 0
Q4_13 0.59 0.29 0.08 0.02 0.02 0
Q4_14 0.11 0.27 0.50 0.09 0.03 0
Q4_15 0.16 0.41 0.33 0.07 0.02 0
Q4_16 0.17 0.33 0.40 0.08 0.02 0
Q4_17 0.26 0.39 0.25 0.08 0.02 0
Q4_18 0.13 0.47 0.34 0.06 0.01 0
Q4_19 0.14 0.35 0.39 0.08 0.04 0Initial Student Centered Items
Item Analayis
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q5_",1:12)])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q5_1 | 0.951 | 0.700 | 0.636 | 2.637 | 0.665 | 0.604 |
| Q5_2 | 1.035 | 0.742 | 0.680 | 2.954 | 0.767 | 0.703 |
| Q5_3 | 1.046 | 0.770 | 0.713 | 2.662 | 0.804 | 0.744 |
| Q5_4 | 1.106 | 0.732 | 0.663 | 3.500 | 0.808 | 0.732 |
| Q5_5 | 1.034 | 0.680 | 0.607 | 3.530 | 0.702 | 0.627 |
| Q5_6 | 0.906 | 0.809 | 0.768 | 2.927 | 0.732 | 0.694 |
| Q5_7 | 0.942 | 0.672 | 0.605 | 2.704 | 0.632 | 0.569 |
| Q5_8 | 1.055 | 0.800 | 0.748 | 2.905 | 0.842 | 0.788 |
| Q5_9 | 0.995 | 0.779 | 0.726 | 2.610 | 0.774 | 0.722 |
| Q5_10 | 0.837 | 0.667 | 0.607 | 2.622 | 0.557 | 0.508 |
| Q5_11 | 0.951 | 0.665 | 0.596 | 2.707 | 0.631 | 0.566 |
| Q5_12 | 0.952 | 0.708 | 0.646 | 2.881 | 0.673 | 0.614 |
out.dat[20:31, 3] <- fit[, 3]Preliminary Reliability
# preliminary reliability
psych::alpha(dat[, paste0("Q5_",1:12)])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q5_", 1:12)])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.92 0.92 0.93 0.49 11 0.0066 2.9 0.72 0.48
lower alpha upper 95% confidence boundaries
0.91 0.92 0.93
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q5_1 0.91 0.91 0.92 0.49 10.6 0.0071 0.0087 0.48
Q5_2 0.91 0.91 0.92 0.48 10.3 0.0073 0.0085 0.48
Q5_3 0.91 0.91 0.92 0.48 10.1 0.0074 0.0081 0.47
Q5_4 0.91 0.91 0.92 0.49 10.4 0.0071 0.0069 0.49
Q5_5 0.91 0.92 0.92 0.49 10.8 0.0070 0.0056 0.49
Q5_6 0.91 0.91 0.92 0.47 9.9 0.0075 0.0078 0.47
Q5_7 0.91 0.91 0.92 0.49 10.7 0.0070 0.0083 0.49
Q5_8 0.91 0.91 0.92 0.48 10.0 0.0075 0.0083 0.47
Q5_9 0.91 0.91 0.92 0.48 10.0 0.0074 0.0084 0.47
Q5_10 0.91 0.91 0.92 0.49 10.7 0.0070 0.0086 0.49
Q5_11 0.91 0.92 0.92 0.50 10.8 0.0070 0.0085 0.49
Q5_12 0.91 0.91 0.92 0.49 10.5 0.0071 0.0094 0.48
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q5_1 328 0.70 0.70 0.67 0.64 2.6 0.95
Q5_2 328 0.74 0.74 0.71 0.68 3.0 1.04
Q5_3 328 0.77 0.77 0.75 0.71 2.7 1.05
Q5_4 328 0.73 0.72 0.71 0.66 3.5 1.11
Q5_5 328 0.68 0.67 0.66 0.61 3.5 1.03
Q5_6 328 0.81 0.81 0.80 0.77 2.9 0.91
Q5_7 328 0.67 0.68 0.64 0.60 2.7 0.94
Q5_8 328 0.80 0.80 0.78 0.75 2.9 1.06
Q5_9 328 0.78 0.78 0.76 0.73 2.6 1.00
Q5_10 328 0.67 0.68 0.64 0.61 2.6 0.84
Q5_11 328 0.66 0.67 0.63 0.60 2.7 0.95
Q5_12 328 0.71 0.71 0.67 0.65 2.9 0.95
Non missing response frequency for each item
1 2 3 4 5 miss
Q5_1 0.12 0.32 0.40 0.14 0.03 0
Q5_2 0.10 0.20 0.38 0.27 0.05 0
Q5_3 0.15 0.29 0.35 0.17 0.04 0
Q5_4 0.08 0.11 0.21 0.46 0.15 0
Q5_5 0.05 0.12 0.21 0.48 0.14 0
Q5_6 0.09 0.15 0.52 0.21 0.02 0
Q5_7 0.13 0.22 0.51 0.11 0.03 0
Q5_8 0.12 0.19 0.39 0.24 0.05 0
Q5_9 0.14 0.32 0.37 0.14 0.03 0
Q5_10 0.13 0.21 0.59 0.06 0.02 0
Q5_11 0.11 0.30 0.41 0.17 0.02 0
Q5_12 0.10 0.17 0.50 0.19 0.04 0Initial Interactive Items
Item Analysis
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q6_",1:11)])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q6_1 | 0.844 | 0.722 | 0.660 | 1.701 | 0.608 | 0.556 |
| Q6_2 | 0.912 | 0.780 | 0.725 | 2.079 | 0.711 | 0.660 |
| Q6_3 | 0.924 | 0.767 | 0.708 | 1.930 | 0.708 | 0.653 |
| Q6_4 | 0.927 | 0.723 | 0.654 | 2.076 | 0.669 | 0.606 |
| Q6_5 | 1.033 | 0.590 | 0.487 | 2.351 | 0.608 | 0.502 |
| Q6_6 | 0.820 | 0.797 | 0.751 | 1.832 | 0.653 | 0.615 |
| Q6_7 | 0.875 | 0.832 | 0.790 | 2.162 | 0.727 | 0.690 |
| Q6_8 | 0.903 | 0.818 | 0.770 | 2.113 | 0.738 | 0.695 |
| Q6_9 | 1.222 | 0.581 | 0.455 | 2.768 | 0.709 | 0.556 |
| Q6_10 | 0.913 | 0.625 | 0.541 | 2.887 | 0.570 | 0.493 |
| Q6_11 | 0.970 | 0.706 | 0.630 | 2.814 | 0.684 | 0.610 |
out.dat[32:42, 3] <- fit[, 3]Preliminary Reliability
# preliminary reliability
psych::alpha(dat[, paste0("Q6_",1:11)])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q6_", 1:11)])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.9 0.91 0.92 0.47 9.9 0.0082 2.2 0.67 0.44
lower alpha upper 95% confidence boundaries
0.89 0.9 0.92
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q6_1 0.89 0.90 0.91 0.47 9.0 0.0091 0.022 0.43
Q6_2 0.89 0.90 0.91 0.46 8.6 0.0093 0.020 0.43
Q6_3 0.89 0.90 0.91 0.46 8.7 0.0093 0.020 0.43
Q6_4 0.89 0.90 0.91 0.47 9.0 0.0090 0.020 0.43
Q6_5 0.90 0.91 0.92 0.50 10.0 0.0082 0.023 0.56
Q6_6 0.89 0.89 0.91 0.46 8.5 0.0094 0.023 0.42
Q6_7 0.89 0.89 0.91 0.45 8.3 0.0097 0.023 0.42
Q6_8 0.89 0.89 0.91 0.46 8.4 0.0096 0.023 0.42
Q6_9 0.91 0.91 0.92 0.51 10.2 0.0077 0.020 0.56
Q6_10 0.90 0.91 0.92 0.49 9.7 0.0086 0.024 0.53
Q6_11 0.89 0.90 0.91 0.48 9.2 0.0090 0.026 0.44
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q6_1 328 0.72 0.73 0.71 0.66 1.7 0.84
Q6_2 328 0.78 0.79 0.78 0.72 2.1 0.91
Q6_3 328 0.77 0.78 0.77 0.71 1.9 0.92
Q6_4 328 0.72 0.74 0.72 0.65 2.1 0.93
Q6_5 328 0.59 0.58 0.51 0.49 2.4 1.03
Q6_6 328 0.80 0.81 0.79 0.75 1.8 0.82
Q6_7 328 0.83 0.84 0.83 0.79 2.2 0.88
Q6_8 328 0.82 0.82 0.81 0.77 2.1 0.90
Q6_9 328 0.58 0.55 0.49 0.46 2.8 1.22
Q6_10 328 0.63 0.62 0.57 0.54 2.9 0.91
Q6_11 328 0.71 0.70 0.66 0.63 2.8 0.97
Non missing response frequency for each item
1 2 3 4 5 miss
Q6_1 0.49 0.36 0.09 0.05 0.00 0
Q6_2 0.27 0.47 0.17 0.07 0.01 0
Q6_3 0.36 0.45 0.13 0.05 0.02 0
Q6_4 0.29 0.43 0.20 0.06 0.01 0
Q6_5 0.19 0.46 0.21 0.10 0.04 0
Q6_6 0.37 0.48 0.11 0.03 0.01 0
Q6_7 0.21 0.50 0.20 0.07 0.01 0
Q6_8 0.25 0.47 0.20 0.07 0.01 0
Q6_9 0.16 0.28 0.30 0.14 0.12 0
Q6_10 0.10 0.15 0.57 0.15 0.04 0
Q6_11 0.11 0.21 0.49 0.14 0.05 0Initial Engagement Items
Item Analysis
fit <- psychometric::item.exam(dat[, paste0("Q7_",1:15)])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q7_1 | 0.901 | 0.748 | 0.703 | 2.463 | 0.673 | 0.633 |
| Q7_2 | 0.857 | 0.738 | 0.694 | 2.692 | 0.631 | 0.594 |
| Q7_3 | 1.012 | 0.712 | 0.656 | 3.558 | 0.720 | 0.663 |
| Q7_4 | 0.980 | 0.732 | 0.681 | 2.841 | 0.716 | 0.666 |
| Q7_5 | 0.948 | 0.808 | 0.770 | 2.875 | 0.765 | 0.729 |
| Q7_6 | 0.920 | 0.745 | 0.699 | 2.933 | 0.684 | 0.642 |
| Q7_7 | 1.014 | 0.736 | 0.683 | 3.652 | 0.745 | 0.692 |
| Q7_8 | 0.877 | 0.731 | 0.685 | 2.814 | 0.640 | 0.600 |
| Q7_9 | 0.993 | 0.716 | 0.662 | 2.567 | 0.710 | 0.656 |
| Q7_10 | 0.945 | 0.686 | 0.631 | 2.280 | 0.648 | 0.595 |
| Q7_11 | 0.840 | 0.745 | 0.704 | 2.744 | 0.625 | 0.590 |
| Q7_12 | 1.023 | 0.674 | 0.611 | 3.390 | 0.688 | 0.624 |
| Q7_13 | 0.989 | 0.454 | 0.370 | 3.579 | 0.449 | 0.365 |
| Q7_14 | 0.986 | 0.605 | 0.535 | 3.610 | 0.595 | 0.527 |
| Q7_15 | 0.905 | 0.665 | 0.609 | 2.509 | 0.601 | 0.550 |
out.dat[43:57, 3] <- fit[, 3]Preliminary Reliability
# preliminary reliability
psych::alpha(dat[, paste0("Q7_",1:15)])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q7_", 1:15)])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.92 0.93 0.94 0.45 12 0.0062 3 0.66 0.47
lower alpha upper 95% confidence boundaries
0.91 0.92 0.94
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q7_1 0.92 0.92 0.93 0.45 11 0.0067 0.017 0.47
Q7_2 0.92 0.92 0.94 0.45 11 0.0067 0.018 0.47
Q7_3 0.92 0.92 0.93 0.45 12 0.0066 0.017 0.48
Q7_4 0.92 0.92 0.94 0.45 11 0.0067 0.017 0.47
Q7_5 0.92 0.92 0.93 0.44 11 0.0069 0.017 0.46
Q7_6 0.92 0.92 0.94 0.45 11 0.0067 0.018 0.46
Q7_7 0.92 0.92 0.93 0.45 11 0.0067 0.017 0.48
Q7_8 0.92 0.92 0.94 0.45 11 0.0067 0.018 0.47
Q7_9 0.92 0.92 0.94 0.45 12 0.0066 0.017 0.47
Q7_10 0.92 0.92 0.93 0.45 12 0.0065 0.015 0.47
Q7_11 0.92 0.92 0.93 0.45 11 0.0067 0.018 0.47
Q7_12 0.92 0.92 0.94 0.46 12 0.0065 0.019 0.49
Q7_13 0.93 0.93 0.94 0.48 13 0.0059 0.011 0.50
Q7_14 0.92 0.92 0.94 0.47 12 0.0063 0.017 0.49
Q7_15 0.92 0.92 0.94 0.46 12 0.0065 0.016 0.48
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q7_1 328 0.75 0.75 0.74 0.70 2.5 0.90
Q7_2 328 0.74 0.74 0.73 0.69 2.7 0.86
Q7_3 328 0.71 0.71 0.69 0.66 3.6 1.01
Q7_4 328 0.73 0.73 0.71 0.68 2.8 0.98
Q7_5 328 0.81 0.81 0.80 0.77 2.9 0.95
Q7_6 328 0.74 0.75 0.72 0.70 2.9 0.92
Q7_7 328 0.74 0.73 0.72 0.68 3.7 1.01
Q7_8 328 0.73 0.73 0.71 0.69 2.8 0.88
Q7_9 328 0.72 0.72 0.70 0.66 2.6 0.99
Q7_10 328 0.69 0.69 0.68 0.63 2.3 0.95
Q7_11 328 0.75 0.75 0.74 0.70 2.7 0.84
Q7_12 328 0.67 0.67 0.64 0.61 3.4 1.02
Q7_13 328 0.45 0.45 0.39 0.37 3.6 0.99
Q7_14 328 0.60 0.60 0.56 0.54 3.6 0.99
Q7_15 328 0.66 0.67 0.65 0.61 2.5 0.91
Non missing response frequency for each item
1 2 3 4 5 miss
Q7_1 0.15 0.35 0.40 0.09 0.01 0
Q7_2 0.11 0.21 0.57 0.09 0.02 0
Q7_3 0.05 0.10 0.22 0.50 0.13 0
Q7_4 0.10 0.21 0.47 0.17 0.05 0
Q7_5 0.10 0.18 0.51 0.17 0.04 0
Q7_6 0.08 0.17 0.53 0.18 0.04 0
Q7_7 0.05 0.09 0.17 0.53 0.16 0
Q7_8 0.10 0.17 0.55 0.15 0.02 0
Q7_9 0.16 0.29 0.41 0.12 0.03 0
Q7_10 0.20 0.45 0.25 0.08 0.02 0
Q7_11 0.10 0.19 0.59 0.11 0.02 0
Q7_12 0.07 0.09 0.32 0.43 0.10 0
Q7_13 0.02 0.11 0.32 0.36 0.19 0
Q7_14 0.05 0.08 0.24 0.48 0.15 0
Q7_15 0.14 0.33 0.43 0.08 0.02 0EFA
Correlations
First, we investigated the raw correlations among the variables.
# Pearson correlation matrix
corr.ppm <- cor(dat)
kable(corr.ppm,digits=3,format = "html")%>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="500px")| Q4_1 | Q4_2 | Q4_3 | Q4_4 | Q4_5 | Q4_6 | Q4_7 | Q4_8 | Q4_9 | Q4_10 | Q4_11 | Q4_12 | Q4_13 | Q4_14 | Q4_15 | Q4_16 | Q4_17 | Q4_18 | Q4_19 | Q5_1 | Q5_2 | Q5_3 | Q5_4 | Q5_5 | Q5_6 | Q5_7 | Q5_8 | Q5_9 | Q5_10 | Q5_11 | Q5_12 | Q6_1 | Q6_2 | Q6_3 | Q6_4 | Q6_5 | Q6_6 | Q6_7 | Q6_8 | Q6_9 | Q6_10 | Q6_11 | Q7_1 | Q7_2 | Q7_3 | Q7_4 | Q7_5 | Q7_6 | Q7_7 | Q7_8 | Q7_9 | Q7_10 | Q7_11 | Q7_12 | Q7_13 | Q7_14 | Q7_15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q4_1 | 1.000 | 0.629 | 0.685 | 0.672 | 0.508 | 0.473 | 0.316 | 0.585 | 0.431 | 0.568 | 0.586 | 0.408 | 0.408 | 0.543 | 0.533 | 0.540 | 0.469 | 0.655 | 0.495 | 0.422 | 0.337 | 0.362 | 0.349 | 0.283 | 0.394 | 0.298 | 0.319 | 0.432 | 0.350 | 0.424 | 0.330 | 0.293 | 0.275 | 0.235 | 0.257 | 0.250 | 0.290 | 0.369 | 0.312 | 0.245 | 0.309 | 0.357 | 0.458 | 0.340 | 0.247 | 0.331 | 0.398 | 0.357 | 0.287 | 0.298 | 0.527 | 0.495 | 0.423 | 0.307 | 0.127 | 0.218 | 0.445 |
| Q4_2 | 0.629 | 1.000 | 0.646 | 0.636 | 0.657 | 0.486 | 0.341 | 0.621 | 0.514 | 0.552 | 0.563 | 0.488 | 0.518 | 0.485 | 0.592 | 0.501 | 0.523 | 0.730 | 0.561 | 0.464 | 0.381 | 0.414 | 0.334 | 0.292 | 0.413 | 0.307 | 0.369 | 0.446 | 0.378 | 0.472 | 0.368 | 0.265 | 0.315 | 0.271 | 0.243 | 0.340 | 0.390 | 0.392 | 0.300 | 0.266 | 0.365 | 0.422 | 0.483 | 0.387 | 0.285 | 0.342 | 0.474 | 0.384 | 0.352 | 0.394 | 0.471 | 0.505 | 0.407 | 0.281 | 0.129 | 0.197 | 0.454 |
| Q4_3 | 0.685 | 0.646 | 1.000 | 0.680 | 0.490 | 0.413 | 0.326 | 0.607 | 0.508 | 0.597 | 0.572 | 0.351 | 0.370 | 0.459 | 0.540 | 0.512 | 0.476 | 0.622 | 0.577 | 0.431 | 0.347 | 0.356 | 0.331 | 0.293 | 0.429 | 0.308 | 0.350 | 0.448 | 0.335 | 0.480 | 0.292 | 0.302 | 0.270 | 0.247 | 0.244 | 0.296 | 0.302 | 0.367 | 0.309 | 0.245 | 0.284 | 0.369 | 0.449 | 0.356 | 0.289 | 0.303 | 0.406 | 0.390 | 0.377 | 0.337 | 0.488 | 0.479 | 0.472 | 0.345 | 0.148 | 0.211 | 0.411 |
| Q4_4 | 0.672 | 0.636 | 0.680 | 1.000 | 0.519 | 0.489 | 0.430 | 0.610 | 0.498 | 0.645 | 0.573 | 0.439 | 0.381 | 0.489 | 0.524 | 0.526 | 0.507 | 0.663 | 0.530 | 0.419 | 0.368 | 0.348 | 0.383 | 0.365 | 0.473 | 0.406 | 0.387 | 0.429 | 0.409 | 0.384 | 0.437 | 0.234 | 0.306 | 0.299 | 0.226 | 0.281 | 0.408 | 0.394 | 0.354 | 0.290 | 0.364 | 0.460 | 0.469 | 0.476 | 0.370 | 0.487 | 0.499 | 0.473 | 0.403 | 0.436 | 0.521 | 0.516 | 0.517 | 0.334 | 0.134 | 0.308 | 0.477 |
| Q4_5 | 0.508 | 0.657 | 0.490 | 0.519 | 1.000 | 0.522 | 0.318 | 0.532 | 0.429 | 0.501 | 0.540 | 0.504 | 0.526 | 0.462 | 0.552 | 0.454 | 0.505 | 0.617 | 0.545 | 0.450 | 0.392 | 0.440 | 0.353 | 0.270 | 0.386 | 0.373 | 0.411 | 0.534 | 0.389 | 0.499 | 0.407 | 0.327 | 0.368 | 0.366 | 0.277 | 0.270 | 0.456 | 0.425 | 0.389 | 0.288 | 0.368 | 0.460 | 0.510 | 0.387 | 0.279 | 0.368 | 0.418 | 0.367 | 0.368 | 0.370 | 0.491 | 0.550 | 0.347 | 0.282 | 0.085 | 0.173 | 0.474 |
| Q4_6 | 0.473 | 0.486 | 0.413 | 0.489 | 0.522 | 1.000 | 0.570 | 0.420 | 0.336 | 0.468 | 0.451 | 0.389 | 0.317 | 0.407 | 0.451 | 0.461 | 0.395 | 0.497 | 0.446 | 0.373 | 0.479 | 0.441 | 0.395 | 0.326 | 0.458 | 0.379 | 0.440 | 0.494 | 0.347 | 0.415 | 0.426 | 0.316 | 0.435 | 0.364 | 0.300 | 0.280 | 0.350 | 0.363 | 0.324 | 0.276 | 0.387 | 0.486 | 0.496 | 0.415 | 0.380 | 0.418 | 0.400 | 0.394 | 0.370 | 0.328 | 0.448 | 0.497 | 0.376 | 0.300 | 0.225 | 0.400 | 0.466 |
| Q4_7 | 0.316 | 0.341 | 0.326 | 0.430 | 0.318 | 0.570 | 1.000 | 0.310 | 0.270 | 0.412 | 0.415 | 0.300 | 0.150 | 0.351 | 0.286 | 0.352 | 0.284 | 0.318 | 0.368 | 0.337 | 0.399 | 0.311 | 0.411 | 0.317 | 0.342 | 0.368 | 0.414 | 0.355 | 0.344 | 0.335 | 0.396 | 0.205 | 0.271 | 0.231 | 0.255 | 0.161 | 0.294 | 0.295 | 0.287 | 0.238 | 0.337 | 0.429 | 0.342 | 0.414 | 0.452 | 0.400 | 0.409 | 0.357 | 0.425 | 0.340 | 0.334 | 0.312 | 0.322 | 0.266 | 0.199 | 0.336 | 0.284 |
| Q4_8 | 0.585 | 0.621 | 0.607 | 0.610 | 0.532 | 0.420 | 0.310 | 1.000 | 0.594 | 0.574 | 0.574 | 0.498 | 0.486 | 0.513 | 0.586 | 0.470 | 0.559 | 0.642 | 0.619 | 0.417 | 0.352 | 0.400 | 0.271 | 0.217 | 0.360 | 0.307 | 0.375 | 0.433 | 0.325 | 0.404 | 0.354 | 0.286 | 0.320 | 0.352 | 0.302 | 0.291 | 0.409 | 0.489 | 0.407 | 0.310 | 0.352 | 0.406 | 0.415 | 0.369 | 0.253 | 0.296 | 0.371 | 0.363 | 0.269 | 0.359 | 0.434 | 0.487 | 0.397 | 0.246 | 0.082 | 0.156 | 0.398 |
| Q4_9 | 0.431 | 0.514 | 0.508 | 0.498 | 0.429 | 0.336 | 0.270 | 0.594 | 1.000 | 0.467 | 0.541 | 0.466 | 0.456 | 0.451 | 0.537 | 0.421 | 0.488 | 0.547 | 0.706 | 0.387 | 0.346 | 0.370 | 0.236 | 0.242 | 0.366 | 0.283 | 0.321 | 0.330 | 0.263 | 0.401 | 0.399 | 0.250 | 0.300 | 0.245 | 0.280 | 0.249 | 0.332 | 0.411 | 0.332 | 0.331 | 0.280 | 0.308 | 0.379 | 0.382 | 0.301 | 0.326 | 0.413 | 0.450 | 0.347 | 0.375 | 0.409 | 0.457 | 0.463 | 0.320 | 0.129 | 0.198 | 0.341 |
| Q4_10 | 0.568 | 0.552 | 0.597 | 0.645 | 0.501 | 0.468 | 0.412 | 0.574 | 0.467 | 1.000 | 0.570 | 0.423 | 0.342 | 0.443 | 0.530 | 0.493 | 0.471 | 0.618 | 0.553 | 0.441 | 0.298 | 0.299 | 0.331 | 0.332 | 0.381 | 0.388 | 0.367 | 0.469 | 0.380 | 0.399 | 0.373 | 0.257 | 0.288 | 0.333 | 0.265 | 0.248 | 0.383 | 0.434 | 0.336 | 0.311 | 0.286 | 0.441 | 0.419 | 0.497 | 0.377 | 0.393 | 0.486 | 0.391 | 0.395 | 0.409 | 0.385 | 0.432 | 0.484 | 0.309 | 0.158 | 0.316 | 0.440 |
| Q4_11 | 0.586 | 0.563 | 0.572 | 0.573 | 0.540 | 0.451 | 0.415 | 0.574 | 0.541 | 0.570 | 1.000 | 0.558 | 0.473 | 0.544 | 0.592 | 0.558 | 0.530 | 0.650 | 0.677 | 0.470 | 0.401 | 0.406 | 0.316 | 0.266 | 0.428 | 0.440 | 0.414 | 0.528 | 0.486 | 0.458 | 0.395 | 0.323 | 0.344 | 0.324 | 0.323 | 0.355 | 0.428 | 0.448 | 0.442 | 0.383 | 0.374 | 0.451 | 0.526 | 0.447 | 0.334 | 0.392 | 0.542 | 0.453 | 0.347 | 0.414 | 0.435 | 0.481 | 0.469 | 0.385 | 0.204 | 0.292 | 0.508 |
| Q4_12 | 0.408 | 0.488 | 0.351 | 0.439 | 0.504 | 0.389 | 0.300 | 0.498 | 0.466 | 0.423 | 0.558 | 1.000 | 0.694 | 0.523 | 0.504 | 0.420 | 0.499 | 0.551 | 0.541 | 0.395 | 0.379 | 0.430 | 0.237 | 0.172 | 0.314 | 0.423 | 0.361 | 0.421 | 0.379 | 0.360 | 0.350 | 0.428 | 0.437 | 0.423 | 0.358 | 0.342 | 0.499 | 0.527 | 0.514 | 0.402 | 0.335 | 0.476 | 0.450 | 0.440 | 0.303 | 0.357 | 0.451 | 0.428 | 0.310 | 0.368 | 0.397 | 0.397 | 0.275 | 0.259 | 0.103 | 0.180 | 0.371 |
| Q4_13 | 0.408 | 0.518 | 0.370 | 0.381 | 0.526 | 0.317 | 0.150 | 0.486 | 0.456 | 0.342 | 0.473 | 0.694 | 1.000 | 0.478 | 0.491 | 0.367 | 0.454 | 0.580 | 0.501 | 0.399 | 0.337 | 0.404 | 0.194 | 0.147 | 0.278 | 0.384 | 0.273 | 0.397 | 0.353 | 0.367 | 0.334 | 0.440 | 0.408 | 0.388 | 0.336 | 0.347 | 0.476 | 0.445 | 0.425 | 0.329 | 0.256 | 0.357 | 0.413 | 0.371 | 0.170 | 0.224 | 0.281 | 0.357 | 0.202 | 0.284 | 0.388 | 0.455 | 0.257 | 0.266 | 0.074 | 0.096 | 0.315 |
| Q4_14 | 0.543 | 0.485 | 0.459 | 0.489 | 0.462 | 0.407 | 0.351 | 0.513 | 0.451 | 0.443 | 0.544 | 0.523 | 0.478 | 1.000 | 0.671 | 0.586 | 0.444 | 0.566 | 0.521 | 0.465 | 0.429 | 0.461 | 0.309 | 0.259 | 0.460 | 0.399 | 0.374 | 0.502 | 0.358 | 0.457 | 0.419 | 0.403 | 0.359 | 0.344 | 0.349 | 0.335 | 0.422 | 0.481 | 0.438 | 0.442 | 0.428 | 0.529 | 0.478 | 0.437 | 0.336 | 0.332 | 0.462 | 0.463 | 0.371 | 0.398 | 0.490 | 0.421 | 0.380 | 0.360 | 0.217 | 0.271 | 0.427 |
| Q4_15 | 0.533 | 0.592 | 0.540 | 0.524 | 0.552 | 0.451 | 0.286 | 0.586 | 0.537 | 0.530 | 0.592 | 0.504 | 0.491 | 0.671 | 1.000 | 0.701 | 0.607 | 0.655 | 0.593 | 0.472 | 0.405 | 0.455 | 0.348 | 0.283 | 0.486 | 0.444 | 0.426 | 0.529 | 0.440 | 0.466 | 0.454 | 0.355 | 0.348 | 0.338 | 0.333 | 0.310 | 0.430 | 0.481 | 0.444 | 0.381 | 0.390 | 0.450 | 0.503 | 0.455 | 0.265 | 0.332 | 0.473 | 0.451 | 0.331 | 0.429 | 0.448 | 0.492 | 0.487 | 0.326 | 0.137 | 0.200 | 0.447 |
| Q4_16 | 0.540 | 0.501 | 0.512 | 0.526 | 0.454 | 0.461 | 0.352 | 0.470 | 0.421 | 0.493 | 0.558 | 0.420 | 0.367 | 0.586 | 0.701 | 1.000 | 0.598 | 0.573 | 0.532 | 0.412 | 0.405 | 0.466 | 0.350 | 0.325 | 0.488 | 0.415 | 0.398 | 0.495 | 0.403 | 0.444 | 0.426 | 0.404 | 0.421 | 0.364 | 0.356 | 0.203 | 0.377 | 0.441 | 0.382 | 0.276 | 0.359 | 0.388 | 0.467 | 0.373 | 0.235 | 0.291 | 0.383 | 0.386 | 0.260 | 0.329 | 0.432 | 0.440 | 0.385 | 0.256 | 0.102 | 0.163 | 0.422 |
| Q4_17 | 0.469 | 0.523 | 0.476 | 0.507 | 0.505 | 0.395 | 0.284 | 0.559 | 0.488 | 0.471 | 0.530 | 0.499 | 0.454 | 0.444 | 0.607 | 0.598 | 1.000 | 0.615 | 0.535 | 0.382 | 0.328 | 0.373 | 0.234 | 0.164 | 0.367 | 0.364 | 0.310 | 0.348 | 0.400 | 0.400 | 0.336 | 0.321 | 0.309 | 0.352 | 0.305 | 0.274 | 0.421 | 0.455 | 0.413 | 0.296 | 0.380 | 0.360 | 0.376 | 0.341 | 0.281 | 0.360 | 0.432 | 0.367 | 0.333 | 0.394 | 0.432 | 0.452 | 0.403 | 0.272 | 0.097 | 0.149 | 0.399 |
| Q4_18 | 0.655 | 0.730 | 0.622 | 0.663 | 0.617 | 0.497 | 0.318 | 0.642 | 0.547 | 0.618 | 0.650 | 0.551 | 0.580 | 0.566 | 0.655 | 0.573 | 0.615 | 1.000 | 0.659 | 0.480 | 0.425 | 0.419 | 0.319 | 0.318 | 0.515 | 0.394 | 0.434 | 0.530 | 0.500 | 0.451 | 0.426 | 0.296 | 0.308 | 0.323 | 0.277 | 0.326 | 0.389 | 0.484 | 0.394 | 0.332 | 0.367 | 0.471 | 0.496 | 0.448 | 0.305 | 0.386 | 0.446 | 0.412 | 0.352 | 0.415 | 0.479 | 0.550 | 0.470 | 0.272 | 0.096 | 0.271 | 0.513 |
| Q4_19 | 0.495 | 0.561 | 0.577 | 0.530 | 0.545 | 0.446 | 0.368 | 0.619 | 0.706 | 0.553 | 0.677 | 0.541 | 0.501 | 0.521 | 0.593 | 0.532 | 0.535 | 0.659 | 1.000 | 0.469 | 0.451 | 0.471 | 0.325 | 0.276 | 0.477 | 0.404 | 0.424 | 0.521 | 0.401 | 0.471 | 0.445 | 0.322 | 0.349 | 0.326 | 0.331 | 0.268 | 0.423 | 0.462 | 0.401 | 0.388 | 0.408 | 0.452 | 0.495 | 0.452 | 0.376 | 0.387 | 0.503 | 0.483 | 0.429 | 0.479 | 0.486 | 0.507 | 0.479 | 0.369 | 0.130 | 0.248 | 0.458 |
| Q5_1 | 0.422 | 0.464 | 0.431 | 0.419 | 0.450 | 0.373 | 0.337 | 0.417 | 0.387 | 0.441 | 0.470 | 0.395 | 0.399 | 0.465 | 0.472 | 0.412 | 0.382 | 0.480 | 0.469 | 1.000 | 0.511 | 0.574 | 0.391 | 0.330 | 0.494 | 0.388 | 0.462 | 0.551 | 0.465 | 0.508 | 0.466 | 0.303 | 0.277 | 0.242 | 0.208 | 0.298 | 0.349 | 0.379 | 0.304 | 0.362 | 0.301 | 0.417 | 0.536 | 0.426 | 0.309 | 0.329 | 0.468 | 0.399 | 0.363 | 0.384 | 0.490 | 0.423 | 0.462 | 0.382 | 0.156 | 0.256 | 0.435 |
| Q5_2 | 0.337 | 0.381 | 0.347 | 0.368 | 0.392 | 0.479 | 0.399 | 0.352 | 0.346 | 0.298 | 0.401 | 0.379 | 0.337 | 0.429 | 0.405 | 0.405 | 0.328 | 0.425 | 0.451 | 0.511 | 1.000 | 0.650 | 0.482 | 0.431 | 0.610 | 0.409 | 0.548 | 0.469 | 0.449 | 0.375 | 0.513 | 0.307 | 0.347 | 0.316 | 0.357 | 0.258 | 0.326 | 0.349 | 0.323 | 0.258 | 0.350 | 0.494 | 0.472 | 0.457 | 0.384 | 0.397 | 0.402 | 0.431 | 0.442 | 0.327 | 0.436 | 0.416 | 0.335 | 0.254 | 0.154 | 0.327 | 0.404 |
| Q5_3 | 0.362 | 0.414 | 0.356 | 0.348 | 0.440 | 0.441 | 0.311 | 0.400 | 0.370 | 0.299 | 0.406 | 0.430 | 0.404 | 0.461 | 0.455 | 0.466 | 0.373 | 0.419 | 0.471 | 0.574 | 0.650 | 1.000 | 0.517 | 0.398 | 0.610 | 0.382 | 0.572 | 0.575 | 0.430 | 0.484 | 0.491 | 0.367 | 0.320 | 0.298 | 0.304 | 0.255 | 0.362 | 0.371 | 0.355 | 0.314 | 0.335 | 0.423 | 0.521 | 0.416 | 0.280 | 0.300 | 0.386 | 0.377 | 0.324 | 0.335 | 0.462 | 0.421 | 0.340 | 0.293 | 0.122 | 0.242 | 0.435 |
| Q5_4 | 0.349 | 0.334 | 0.331 | 0.383 | 0.353 | 0.395 | 0.411 | 0.271 | 0.236 | 0.331 | 0.316 | 0.237 | 0.194 | 0.309 | 0.348 | 0.350 | 0.234 | 0.319 | 0.325 | 0.391 | 0.482 | 0.517 | 1.000 | 0.781 | 0.586 | 0.371 | 0.539 | 0.433 | 0.396 | 0.355 | 0.440 | 0.200 | 0.246 | 0.253 | 0.198 | 0.210 | 0.268 | 0.210 | 0.191 | 0.174 | 0.298 | 0.386 | 0.414 | 0.421 | 0.439 | 0.308 | 0.363 | 0.346 | 0.469 | 0.314 | 0.345 | 0.322 | 0.356 | 0.308 | 0.145 | 0.370 | 0.310 |
| Q5_5 | 0.283 | 0.292 | 0.293 | 0.365 | 0.270 | 0.326 | 0.317 | 0.217 | 0.242 | 0.332 | 0.266 | 0.172 | 0.147 | 0.259 | 0.283 | 0.325 | 0.164 | 0.318 | 0.276 | 0.330 | 0.431 | 0.398 | 0.781 | 1.000 | 0.587 | 0.353 | 0.534 | 0.395 | 0.307 | 0.348 | 0.387 | 0.116 | 0.176 | 0.193 | 0.165 | 0.186 | 0.185 | 0.158 | 0.119 | 0.110 | 0.225 | 0.272 | 0.356 | 0.413 | 0.359 | 0.304 | 0.302 | 0.292 | 0.389 | 0.315 | 0.284 | 0.260 | 0.365 | 0.333 | 0.150 | 0.348 | 0.266 |
| Q5_6 | 0.394 | 0.413 | 0.429 | 0.473 | 0.386 | 0.458 | 0.342 | 0.360 | 0.366 | 0.381 | 0.428 | 0.314 | 0.278 | 0.460 | 0.486 | 0.488 | 0.367 | 0.515 | 0.477 | 0.494 | 0.610 | 0.610 | 0.586 | 0.587 | 1.000 | 0.530 | 0.668 | 0.572 | 0.460 | 0.426 | 0.522 | 0.247 | 0.325 | 0.337 | 0.294 | 0.296 | 0.334 | 0.409 | 0.350 | 0.250 | 0.397 | 0.461 | 0.488 | 0.444 | 0.402 | 0.407 | 0.470 | 0.446 | 0.422 | 0.395 | 0.454 | 0.453 | 0.426 | 0.348 | 0.140 | 0.372 | 0.404 |
| Q5_7 | 0.298 | 0.307 | 0.308 | 0.406 | 0.373 | 0.379 | 0.368 | 0.307 | 0.283 | 0.388 | 0.440 | 0.423 | 0.384 | 0.399 | 0.444 | 0.415 | 0.364 | 0.394 | 0.404 | 0.388 | 0.409 | 0.382 | 0.371 | 0.353 | 0.530 | 1.000 | 0.538 | 0.571 | 0.525 | 0.412 | 0.445 | 0.346 | 0.437 | 0.436 | 0.344 | 0.317 | 0.442 | 0.511 | 0.460 | 0.405 | 0.356 | 0.492 | 0.464 | 0.497 | 0.340 | 0.360 | 0.431 | 0.415 | 0.311 | 0.429 | 0.369 | 0.372 | 0.403 | 0.298 | 0.135 | 0.211 | 0.331 |
| Q5_8 | 0.319 | 0.369 | 0.350 | 0.387 | 0.411 | 0.440 | 0.414 | 0.375 | 0.321 | 0.367 | 0.414 | 0.361 | 0.273 | 0.374 | 0.426 | 0.398 | 0.310 | 0.434 | 0.424 | 0.462 | 0.548 | 0.572 | 0.539 | 0.534 | 0.668 | 0.538 | 1.000 | 0.637 | 0.482 | 0.469 | 0.497 | 0.267 | 0.335 | 0.366 | 0.289 | 0.348 | 0.335 | 0.358 | 0.351 | 0.251 | 0.382 | 0.497 | 0.506 | 0.458 | 0.416 | 0.426 | 0.438 | 0.444 | 0.386 | 0.424 | 0.434 | 0.422 | 0.421 | 0.360 | 0.152 | 0.323 | 0.380 |
| Q5_9 | 0.432 | 0.446 | 0.448 | 0.429 | 0.534 | 0.494 | 0.355 | 0.433 | 0.330 | 0.469 | 0.528 | 0.421 | 0.397 | 0.502 | 0.529 | 0.495 | 0.348 | 0.530 | 0.521 | 0.551 | 0.469 | 0.575 | 0.433 | 0.395 | 0.572 | 0.571 | 0.637 | 1.000 | 0.553 | 0.544 | 0.519 | 0.411 | 0.432 | 0.442 | 0.340 | 0.330 | 0.448 | 0.483 | 0.447 | 0.363 | 0.449 | 0.564 | 0.587 | 0.461 | 0.347 | 0.435 | 0.450 | 0.399 | 0.307 | 0.421 | 0.521 | 0.500 | 0.433 | 0.342 | 0.103 | 0.265 | 0.506 |
| Q5_10 | 0.350 | 0.378 | 0.335 | 0.409 | 0.389 | 0.347 | 0.344 | 0.325 | 0.263 | 0.380 | 0.486 | 0.379 | 0.353 | 0.358 | 0.440 | 0.403 | 0.400 | 0.500 | 0.401 | 0.465 | 0.449 | 0.430 | 0.396 | 0.307 | 0.460 | 0.525 | 0.482 | 0.553 | 1.000 | 0.425 | 0.435 | 0.264 | 0.280 | 0.333 | 0.301 | 0.221 | 0.420 | 0.384 | 0.388 | 0.261 | 0.364 | 0.429 | 0.436 | 0.426 | 0.289 | 0.363 | 0.422 | 0.396 | 0.335 | 0.416 | 0.398 | 0.417 | 0.414 | 0.298 | 0.040 | 0.280 | 0.344 |
| Q5_11 | 0.424 | 0.472 | 0.480 | 0.384 | 0.499 | 0.415 | 0.335 | 0.404 | 0.401 | 0.399 | 0.458 | 0.360 | 0.367 | 0.457 | 0.466 | 0.444 | 0.400 | 0.451 | 0.471 | 0.508 | 0.375 | 0.484 | 0.355 | 0.348 | 0.426 | 0.412 | 0.469 | 0.544 | 0.425 | 1.000 | 0.495 | 0.329 | 0.376 | 0.335 | 0.303 | 0.369 | 0.376 | 0.406 | 0.370 | 0.339 | 0.423 | 0.415 | 0.576 | 0.433 | 0.383 | 0.446 | 0.454 | 0.394 | 0.354 | 0.371 | 0.526 | 0.472 | 0.450 | 0.454 | 0.317 | 0.279 | 0.422 |
| Q5_12 | 0.330 | 0.368 | 0.292 | 0.437 | 0.407 | 0.426 | 0.396 | 0.354 | 0.399 | 0.373 | 0.395 | 0.350 | 0.334 | 0.419 | 0.454 | 0.426 | 0.336 | 0.426 | 0.445 | 0.466 | 0.513 | 0.491 | 0.440 | 0.387 | 0.522 | 0.445 | 0.497 | 0.519 | 0.435 | 0.495 | 1.000 | 0.275 | 0.423 | 0.348 | 0.284 | 0.285 | 0.378 | 0.405 | 0.346 | 0.265 | 0.371 | 0.483 | 0.481 | 0.398 | 0.412 | 0.485 | 0.482 | 0.396 | 0.410 | 0.453 | 0.486 | 0.455 | 0.432 | 0.396 | 0.200 | 0.354 | 0.447 |
| Q6_1 | 0.293 | 0.265 | 0.302 | 0.234 | 0.327 | 0.316 | 0.205 | 0.286 | 0.250 | 0.257 | 0.323 | 0.428 | 0.440 | 0.403 | 0.355 | 0.404 | 0.321 | 0.296 | 0.322 | 0.303 | 0.307 | 0.367 | 0.200 | 0.116 | 0.247 | 0.346 | 0.267 | 0.411 | 0.264 | 0.329 | 0.275 | 1.000 | 0.695 | 0.652 | 0.573 | 0.286 | 0.577 | 0.559 | 0.566 | 0.280 | 0.286 | 0.369 | 0.472 | 0.300 | 0.117 | 0.220 | 0.278 | 0.289 | 0.143 | 0.185 | 0.378 | 0.389 | 0.237 | 0.256 | 0.124 | 0.080 | 0.364 |
| Q6_2 | 0.275 | 0.315 | 0.270 | 0.306 | 0.368 | 0.435 | 0.271 | 0.320 | 0.300 | 0.288 | 0.344 | 0.437 | 0.408 | 0.359 | 0.348 | 0.421 | 0.309 | 0.308 | 0.349 | 0.277 | 0.347 | 0.320 | 0.246 | 0.176 | 0.325 | 0.437 | 0.335 | 0.432 | 0.280 | 0.376 | 0.423 | 0.695 | 1.000 | 0.725 | 0.723 | 0.347 | 0.595 | 0.593 | 0.631 | 0.253 | 0.312 | 0.418 | 0.461 | 0.345 | 0.237 | 0.322 | 0.344 | 0.353 | 0.152 | 0.217 | 0.399 | 0.435 | 0.286 | 0.242 | 0.095 | 0.150 | 0.344 |
| Q6_3 | 0.235 | 0.271 | 0.247 | 0.299 | 0.366 | 0.364 | 0.231 | 0.352 | 0.245 | 0.333 | 0.324 | 0.423 | 0.388 | 0.344 | 0.338 | 0.364 | 0.352 | 0.323 | 0.326 | 0.242 | 0.316 | 0.298 | 0.253 | 0.193 | 0.337 | 0.436 | 0.366 | 0.442 | 0.333 | 0.335 | 0.348 | 0.652 | 0.725 | 1.000 | 0.652 | 0.372 | 0.638 | 0.668 | 0.574 | 0.227 | 0.269 | 0.415 | 0.447 | 0.313 | 0.271 | 0.295 | 0.294 | 0.333 | 0.179 | 0.214 | 0.337 | 0.404 | 0.249 | 0.217 | 0.095 | 0.164 | 0.346 |
| Q6_4 | 0.257 | 0.243 | 0.244 | 0.226 | 0.277 | 0.300 | 0.255 | 0.302 | 0.280 | 0.265 | 0.323 | 0.358 | 0.336 | 0.349 | 0.333 | 0.356 | 0.305 | 0.277 | 0.331 | 0.208 | 0.357 | 0.304 | 0.198 | 0.165 | 0.294 | 0.344 | 0.289 | 0.340 | 0.301 | 0.303 | 0.284 | 0.573 | 0.723 | 0.652 | 1.000 | 0.250 | 0.560 | 0.592 | 0.640 | 0.191 | 0.299 | 0.366 | 0.357 | 0.315 | 0.189 | 0.320 | 0.265 | 0.329 | 0.142 | 0.217 | 0.328 | 0.394 | 0.304 | 0.162 | 0.022 | 0.096 | 0.278 |
| Q6_5 | 0.250 | 0.340 | 0.296 | 0.281 | 0.270 | 0.280 | 0.161 | 0.291 | 0.249 | 0.248 | 0.355 | 0.342 | 0.347 | 0.335 | 0.310 | 0.203 | 0.274 | 0.326 | 0.268 | 0.298 | 0.258 | 0.255 | 0.210 | 0.186 | 0.296 | 0.317 | 0.348 | 0.330 | 0.221 | 0.369 | 0.285 | 0.286 | 0.347 | 0.372 | 0.250 | 1.000 | 0.406 | 0.435 | 0.397 | 0.329 | 0.421 | 0.349 | 0.377 | 0.354 | 0.210 | 0.243 | 0.295 | 0.295 | 0.234 | 0.275 | 0.372 | 0.410 | 0.337 | 0.322 | 0.211 | 0.153 | 0.345 |
| Q6_6 | 0.290 | 0.390 | 0.302 | 0.408 | 0.456 | 0.350 | 0.294 | 0.409 | 0.332 | 0.383 | 0.428 | 0.499 | 0.476 | 0.422 | 0.430 | 0.377 | 0.421 | 0.389 | 0.423 | 0.349 | 0.326 | 0.362 | 0.268 | 0.185 | 0.334 | 0.442 | 0.335 | 0.448 | 0.420 | 0.376 | 0.378 | 0.577 | 0.595 | 0.638 | 0.560 | 0.406 | 1.000 | 0.682 | 0.690 | 0.346 | 0.395 | 0.534 | 0.519 | 0.440 | 0.249 | 0.389 | 0.406 | 0.387 | 0.209 | 0.382 | 0.440 | 0.534 | 0.413 | 0.235 | 0.060 | 0.104 | 0.445 |
| Q6_7 | 0.369 | 0.392 | 0.367 | 0.394 | 0.425 | 0.363 | 0.295 | 0.489 | 0.411 | 0.434 | 0.448 | 0.527 | 0.445 | 0.481 | 0.481 | 0.441 | 0.455 | 0.484 | 0.462 | 0.379 | 0.349 | 0.371 | 0.210 | 0.158 | 0.409 | 0.511 | 0.358 | 0.483 | 0.384 | 0.406 | 0.405 | 0.559 | 0.593 | 0.668 | 0.592 | 0.435 | 0.682 | 1.000 | 0.727 | 0.401 | 0.444 | 0.558 | 0.514 | 0.450 | 0.302 | 0.383 | 0.437 | 0.409 | 0.274 | 0.394 | 0.447 | 0.503 | 0.414 | 0.271 | 0.075 | 0.151 | 0.444 |
| Q6_8 | 0.312 | 0.300 | 0.309 | 0.354 | 0.389 | 0.324 | 0.287 | 0.407 | 0.332 | 0.336 | 0.442 | 0.514 | 0.425 | 0.438 | 0.444 | 0.382 | 0.413 | 0.394 | 0.401 | 0.304 | 0.323 | 0.355 | 0.191 | 0.119 | 0.350 | 0.460 | 0.351 | 0.447 | 0.388 | 0.370 | 0.346 | 0.566 | 0.631 | 0.574 | 0.640 | 0.397 | 0.690 | 0.727 | 1.000 | 0.403 | 0.416 | 0.499 | 0.477 | 0.401 | 0.235 | 0.425 | 0.424 | 0.421 | 0.196 | 0.347 | 0.436 | 0.486 | 0.405 | 0.250 | 0.057 | 0.108 | 0.457 |
| Q6_9 | 0.245 | 0.266 | 0.245 | 0.290 | 0.288 | 0.276 | 0.238 | 0.310 | 0.331 | 0.311 | 0.383 | 0.402 | 0.329 | 0.442 | 0.381 | 0.276 | 0.296 | 0.332 | 0.388 | 0.362 | 0.258 | 0.314 | 0.174 | 0.110 | 0.250 | 0.405 | 0.251 | 0.363 | 0.261 | 0.339 | 0.265 | 0.280 | 0.253 | 0.227 | 0.191 | 0.329 | 0.346 | 0.401 | 0.403 | 1.000 | 0.472 | 0.469 | 0.387 | 0.355 | 0.340 | 0.273 | 0.432 | 0.451 | 0.335 | 0.365 | 0.335 | 0.342 | 0.306 | 0.359 | 0.352 | 0.239 | 0.336 |
| Q6_10 | 0.309 | 0.365 | 0.284 | 0.364 | 0.368 | 0.387 | 0.337 | 0.352 | 0.280 | 0.286 | 0.374 | 0.335 | 0.256 | 0.428 | 0.390 | 0.359 | 0.380 | 0.367 | 0.408 | 0.301 | 0.350 | 0.335 | 0.298 | 0.225 | 0.397 | 0.356 | 0.382 | 0.449 | 0.364 | 0.423 | 0.371 | 0.286 | 0.312 | 0.269 | 0.299 | 0.421 | 0.395 | 0.444 | 0.416 | 0.472 | 1.000 | 0.577 | 0.431 | 0.378 | 0.300 | 0.325 | 0.422 | 0.402 | 0.360 | 0.390 | 0.455 | 0.419 | 0.397 | 0.388 | 0.198 | 0.263 | 0.428 |
| Q6_11 | 0.357 | 0.422 | 0.369 | 0.460 | 0.460 | 0.486 | 0.429 | 0.406 | 0.308 | 0.441 | 0.451 | 0.476 | 0.357 | 0.529 | 0.450 | 0.388 | 0.360 | 0.471 | 0.452 | 0.417 | 0.494 | 0.423 | 0.386 | 0.272 | 0.461 | 0.492 | 0.497 | 0.564 | 0.429 | 0.415 | 0.483 | 0.369 | 0.418 | 0.415 | 0.366 | 0.349 | 0.534 | 0.558 | 0.499 | 0.469 | 0.577 | 1.000 | 0.568 | 0.505 | 0.486 | 0.471 | 0.523 | 0.493 | 0.469 | 0.441 | 0.484 | 0.477 | 0.396 | 0.366 | 0.218 | 0.378 | 0.467 |
| Q7_1 | 0.458 | 0.483 | 0.449 | 0.469 | 0.510 | 0.496 | 0.342 | 0.415 | 0.379 | 0.419 | 0.526 | 0.450 | 0.413 | 0.478 | 0.503 | 0.467 | 0.376 | 0.496 | 0.495 | 0.536 | 0.472 | 0.521 | 0.414 | 0.356 | 0.488 | 0.464 | 0.506 | 0.587 | 0.436 | 0.576 | 0.481 | 0.472 | 0.461 | 0.447 | 0.357 | 0.377 | 0.519 | 0.514 | 0.477 | 0.387 | 0.431 | 0.568 | 1.000 | 0.637 | 0.447 | 0.530 | 0.558 | 0.521 | 0.408 | 0.454 | 0.631 | 0.640 | 0.533 | 0.447 | 0.195 | 0.269 | 0.632 |
| Q7_2 | 0.340 | 0.387 | 0.356 | 0.476 | 0.387 | 0.415 | 0.414 | 0.369 | 0.382 | 0.497 | 0.447 | 0.440 | 0.371 | 0.437 | 0.455 | 0.373 | 0.341 | 0.448 | 0.452 | 0.426 | 0.457 | 0.416 | 0.421 | 0.413 | 0.444 | 0.497 | 0.458 | 0.461 | 0.426 | 0.433 | 0.398 | 0.300 | 0.345 | 0.313 | 0.315 | 0.354 | 0.440 | 0.450 | 0.401 | 0.355 | 0.378 | 0.505 | 0.637 | 1.000 | 0.502 | 0.572 | 0.566 | 0.517 | 0.464 | 0.514 | 0.483 | 0.492 | 0.604 | 0.378 | 0.218 | 0.368 | 0.503 |
| Q7_3 | 0.247 | 0.285 | 0.289 | 0.370 | 0.279 | 0.380 | 0.452 | 0.253 | 0.301 | 0.377 | 0.334 | 0.303 | 0.170 | 0.336 | 0.265 | 0.235 | 0.281 | 0.305 | 0.376 | 0.309 | 0.384 | 0.280 | 0.439 | 0.359 | 0.402 | 0.340 | 0.416 | 0.347 | 0.289 | 0.383 | 0.412 | 0.117 | 0.237 | 0.271 | 0.189 | 0.210 | 0.249 | 0.302 | 0.235 | 0.340 | 0.300 | 0.486 | 0.447 | 0.502 | 1.000 | 0.565 | 0.545 | 0.540 | 0.732 | 0.465 | 0.308 | 0.290 | 0.410 | 0.457 | 0.324 | 0.485 | 0.333 |
| Q7_4 | 0.331 | 0.342 | 0.303 | 0.487 | 0.368 | 0.418 | 0.400 | 0.296 | 0.326 | 0.393 | 0.392 | 0.357 | 0.224 | 0.332 | 0.332 | 0.291 | 0.360 | 0.386 | 0.387 | 0.329 | 0.397 | 0.300 | 0.308 | 0.304 | 0.407 | 0.360 | 0.426 | 0.435 | 0.363 | 0.446 | 0.485 | 0.220 | 0.322 | 0.295 | 0.320 | 0.243 | 0.389 | 0.383 | 0.425 | 0.273 | 0.325 | 0.471 | 0.530 | 0.572 | 0.565 | 1.000 | 0.670 | 0.525 | 0.446 | 0.507 | 0.508 | 0.464 | 0.512 | 0.373 | 0.155 | 0.360 | 0.505 |
| Q7_5 | 0.398 | 0.474 | 0.406 | 0.499 | 0.418 | 0.400 | 0.409 | 0.371 | 0.413 | 0.486 | 0.542 | 0.451 | 0.281 | 0.462 | 0.473 | 0.383 | 0.432 | 0.446 | 0.503 | 0.468 | 0.402 | 0.386 | 0.363 | 0.302 | 0.470 | 0.431 | 0.438 | 0.450 | 0.422 | 0.454 | 0.482 | 0.278 | 0.344 | 0.294 | 0.265 | 0.295 | 0.406 | 0.437 | 0.424 | 0.432 | 0.422 | 0.523 | 0.558 | 0.566 | 0.545 | 0.670 | 1.000 | 0.597 | 0.569 | 0.604 | 0.556 | 0.555 | 0.578 | 0.467 | 0.299 | 0.412 | 0.506 |
| Q7_6 | 0.357 | 0.384 | 0.390 | 0.473 | 0.367 | 0.394 | 0.357 | 0.363 | 0.450 | 0.391 | 0.453 | 0.428 | 0.357 | 0.463 | 0.451 | 0.386 | 0.367 | 0.412 | 0.483 | 0.399 | 0.431 | 0.377 | 0.346 | 0.292 | 0.446 | 0.415 | 0.444 | 0.399 | 0.396 | 0.394 | 0.396 | 0.289 | 0.353 | 0.333 | 0.329 | 0.295 | 0.387 | 0.409 | 0.421 | 0.451 | 0.402 | 0.493 | 0.521 | 0.517 | 0.540 | 0.525 | 0.597 | 1.000 | 0.562 | 0.546 | 0.467 | 0.476 | 0.508 | 0.473 | 0.234 | 0.487 | 0.368 |
| Q7_7 | 0.287 | 0.352 | 0.377 | 0.403 | 0.368 | 0.370 | 0.425 | 0.269 | 0.347 | 0.395 | 0.347 | 0.310 | 0.202 | 0.371 | 0.331 | 0.260 | 0.333 | 0.352 | 0.429 | 0.363 | 0.442 | 0.324 | 0.469 | 0.389 | 0.422 | 0.311 | 0.386 | 0.307 | 0.335 | 0.354 | 0.410 | 0.143 | 0.152 | 0.179 | 0.142 | 0.234 | 0.209 | 0.274 | 0.196 | 0.335 | 0.360 | 0.469 | 0.408 | 0.464 | 0.732 | 0.446 | 0.569 | 0.562 | 1.000 | 0.597 | 0.385 | 0.332 | 0.459 | 0.503 | 0.378 | 0.530 | 0.287 |
| Q7_8 | 0.298 | 0.394 | 0.337 | 0.436 | 0.370 | 0.328 | 0.340 | 0.359 | 0.375 | 0.409 | 0.414 | 0.368 | 0.284 | 0.398 | 0.429 | 0.329 | 0.394 | 0.415 | 0.479 | 0.384 | 0.327 | 0.335 | 0.314 | 0.315 | 0.395 | 0.429 | 0.424 | 0.421 | 0.416 | 0.371 | 0.453 | 0.185 | 0.217 | 0.214 | 0.217 | 0.275 | 0.382 | 0.394 | 0.347 | 0.365 | 0.390 | 0.441 | 0.454 | 0.514 | 0.465 | 0.507 | 0.604 | 0.546 | 0.597 | 1.000 | 0.515 | 0.465 | 0.620 | 0.391 | 0.255 | 0.393 | 0.389 |
| Q7_9 | 0.527 | 0.471 | 0.488 | 0.521 | 0.491 | 0.448 | 0.334 | 0.434 | 0.409 | 0.385 | 0.435 | 0.397 | 0.388 | 0.490 | 0.448 | 0.432 | 0.432 | 0.479 | 0.486 | 0.490 | 0.436 | 0.462 | 0.345 | 0.284 | 0.454 | 0.369 | 0.434 | 0.521 | 0.398 | 0.526 | 0.486 | 0.378 | 0.399 | 0.337 | 0.328 | 0.372 | 0.440 | 0.447 | 0.436 | 0.335 | 0.455 | 0.484 | 0.631 | 0.483 | 0.308 | 0.508 | 0.556 | 0.467 | 0.385 | 0.515 | 1.000 | 0.693 | 0.567 | 0.375 | 0.188 | 0.277 | 0.593 |
| Q7_10 | 0.495 | 0.505 | 0.479 | 0.516 | 0.550 | 0.497 | 0.312 | 0.487 | 0.457 | 0.432 | 0.481 | 0.397 | 0.455 | 0.421 | 0.492 | 0.440 | 0.452 | 0.550 | 0.507 | 0.423 | 0.416 | 0.421 | 0.322 | 0.260 | 0.453 | 0.372 | 0.422 | 0.500 | 0.417 | 0.472 | 0.455 | 0.389 | 0.435 | 0.404 | 0.394 | 0.410 | 0.534 | 0.503 | 0.486 | 0.342 | 0.419 | 0.477 | 0.640 | 0.492 | 0.290 | 0.464 | 0.555 | 0.476 | 0.332 | 0.465 | 0.693 | 1.000 | 0.538 | 0.323 | 0.127 | 0.190 | 0.680 |
| Q7_11 | 0.423 | 0.407 | 0.472 | 0.517 | 0.347 | 0.376 | 0.322 | 0.397 | 0.463 | 0.484 | 0.469 | 0.275 | 0.257 | 0.380 | 0.487 | 0.385 | 0.403 | 0.470 | 0.479 | 0.462 | 0.335 | 0.340 | 0.356 | 0.365 | 0.426 | 0.403 | 0.421 | 0.433 | 0.414 | 0.450 | 0.432 | 0.237 | 0.286 | 0.249 | 0.304 | 0.337 | 0.413 | 0.414 | 0.405 | 0.306 | 0.397 | 0.396 | 0.533 | 0.604 | 0.410 | 0.512 | 0.578 | 0.508 | 0.459 | 0.620 | 0.567 | 0.538 | 1.000 | 0.519 | 0.260 | 0.344 | 0.446 |
| Q7_12 | 0.307 | 0.281 | 0.345 | 0.334 | 0.282 | 0.300 | 0.266 | 0.246 | 0.320 | 0.309 | 0.385 | 0.259 | 0.266 | 0.360 | 0.326 | 0.256 | 0.272 | 0.272 | 0.369 | 0.382 | 0.254 | 0.293 | 0.308 | 0.333 | 0.348 | 0.298 | 0.360 | 0.342 | 0.298 | 0.454 | 0.396 | 0.256 | 0.242 | 0.217 | 0.162 | 0.322 | 0.235 | 0.271 | 0.250 | 0.359 | 0.388 | 0.366 | 0.447 | 0.378 | 0.457 | 0.373 | 0.467 | 0.473 | 0.503 | 0.391 | 0.375 | 0.323 | 0.519 | 1.000 | 0.483 | 0.449 | 0.366 |
| Q7_13 | 0.127 | 0.129 | 0.148 | 0.134 | 0.085 | 0.225 | 0.199 | 0.082 | 0.129 | 0.158 | 0.204 | 0.103 | 0.074 | 0.217 | 0.137 | 0.102 | 0.097 | 0.096 | 0.130 | 0.156 | 0.154 | 0.122 | 0.145 | 0.150 | 0.140 | 0.135 | 0.152 | 0.103 | 0.040 | 0.317 | 0.200 | 0.124 | 0.095 | 0.095 | 0.022 | 0.211 | 0.060 | 0.075 | 0.057 | 0.352 | 0.198 | 0.218 | 0.195 | 0.218 | 0.324 | 0.155 | 0.299 | 0.234 | 0.378 | 0.255 | 0.188 | 0.127 | 0.260 | 0.483 | 1.000 | 0.424 | 0.138 |
| Q7_14 | 0.218 | 0.197 | 0.211 | 0.308 | 0.173 | 0.400 | 0.336 | 0.156 | 0.198 | 0.316 | 0.292 | 0.180 | 0.096 | 0.271 | 0.200 | 0.163 | 0.149 | 0.271 | 0.248 | 0.256 | 0.327 | 0.242 | 0.370 | 0.348 | 0.372 | 0.211 | 0.323 | 0.265 | 0.280 | 0.279 | 0.354 | 0.080 | 0.150 | 0.164 | 0.096 | 0.153 | 0.104 | 0.151 | 0.108 | 0.239 | 0.263 | 0.378 | 0.269 | 0.368 | 0.485 | 0.360 | 0.412 | 0.487 | 0.530 | 0.393 | 0.277 | 0.190 | 0.344 | 0.449 | 0.424 | 1.000 | 0.285 |
| Q7_15 | 0.445 | 0.454 | 0.411 | 0.477 | 0.474 | 0.466 | 0.284 | 0.398 | 0.341 | 0.440 | 0.508 | 0.371 | 0.315 | 0.427 | 0.447 | 0.422 | 0.399 | 0.513 | 0.458 | 0.435 | 0.404 | 0.435 | 0.310 | 0.266 | 0.404 | 0.331 | 0.380 | 0.506 | 0.344 | 0.422 | 0.447 | 0.364 | 0.344 | 0.346 | 0.278 | 0.345 | 0.445 | 0.444 | 0.457 | 0.336 | 0.428 | 0.467 | 0.632 | 0.503 | 0.333 | 0.505 | 0.506 | 0.368 | 0.287 | 0.389 | 0.593 | 0.680 | 0.446 | 0.366 | 0.138 | 0.285 | 1.000 |
# Polychoric Correlation
corr.pc <- polychoric(dat)Warning in cor.smooth(mat): Matrix was not positive definite, smoothing was donekable(corr.pc$rho,digits=3,format = "html")%>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="500px")| Q4_1 | Q4_2 | Q4_3 | Q4_4 | Q4_5 | Q4_6 | Q4_7 | Q4_8 | Q4_9 | Q4_10 | Q4_11 | Q4_12 | Q4_13 | Q4_14 | Q4_15 | Q4_16 | Q4_17 | Q4_18 | Q4_19 | Q5_1 | Q5_2 | Q5_3 | Q5_4 | Q5_5 | Q5_6 | Q5_7 | Q5_8 | Q5_9 | Q5_10 | Q5_11 | Q5_12 | Q6_1 | Q6_2 | Q6_3 | Q6_4 | Q6_5 | Q6_6 | Q6_7 | Q6_8 | Q6_9 | Q6_10 | Q6_11 | Q7_1 | Q7_2 | Q7_3 | Q7_4 | Q7_5 | Q7_6 | Q7_7 | Q7_8 | Q7_9 | Q7_10 | Q7_11 | Q7_12 | Q7_13 | Q7_14 | Q7_15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q4_1 | 1.000 | 0.701 | 0.761 | 0.752 | 0.574 | 0.522 | 0.349 | 0.655 | 0.491 | 0.643 | 0.648 | 0.471 | 0.500 | 0.607 | 0.602 | 0.606 | 0.533 | 0.734 | 0.558 | 0.474 | 0.377 | 0.407 | 0.381 | 0.319 | 0.438 | 0.340 | 0.347 | 0.487 | 0.404 | 0.476 | 0.364 | 0.370 | 0.325 | 0.304 | 0.307 | 0.300 | 0.356 | 0.434 | 0.370 | 0.288 | 0.345 | 0.404 | 0.516 | 0.390 | 0.266 | 0.369 | 0.444 | 0.406 | 0.334 | 0.332 | 0.586 | 0.570 | 0.484 | 0.351 | 0.134 | 0.229 | 0.502 |
| Q4_2 | 0.701 | 1.000 | 0.721 | 0.717 | 0.731 | 0.551 | 0.380 | 0.693 | 0.576 | 0.629 | 0.632 | 0.557 | 0.615 | 0.534 | 0.663 | 0.565 | 0.591 | 0.804 | 0.624 | 0.530 | 0.434 | 0.472 | 0.384 | 0.338 | 0.479 | 0.351 | 0.423 | 0.512 | 0.455 | 0.532 | 0.421 | 0.351 | 0.372 | 0.337 | 0.285 | 0.400 | 0.475 | 0.468 | 0.359 | 0.318 | 0.410 | 0.483 | 0.557 | 0.453 | 0.315 | 0.388 | 0.546 | 0.442 | 0.411 | 0.449 | 0.537 | 0.591 | 0.482 | 0.314 | 0.137 | 0.217 | 0.522 |
| Q4_3 | 0.761 | 0.721 | 1.000 | 0.766 | 0.550 | 0.463 | 0.347 | 0.681 | 0.579 | 0.676 | 0.632 | 0.409 | 0.451 | 0.502 | 0.603 | 0.571 | 0.542 | 0.694 | 0.640 | 0.485 | 0.387 | 0.400 | 0.357 | 0.328 | 0.488 | 0.348 | 0.388 | 0.505 | 0.385 | 0.538 | 0.323 | 0.393 | 0.313 | 0.317 | 0.284 | 0.360 | 0.363 | 0.432 | 0.357 | 0.290 | 0.319 | 0.415 | 0.504 | 0.398 | 0.308 | 0.338 | 0.454 | 0.443 | 0.411 | 0.379 | 0.546 | 0.549 | 0.535 | 0.374 | 0.154 | 0.230 | 0.463 |
| Q4_4 | 0.752 | 0.717 | 0.766 | 1.000 | 0.590 | 0.544 | 0.469 | 0.690 | 0.572 | 0.735 | 0.642 | 0.521 | 0.467 | 0.537 | 0.589 | 0.592 | 0.591 | 0.749 | 0.594 | 0.474 | 0.401 | 0.399 | 0.414 | 0.399 | 0.523 | 0.446 | 0.422 | 0.485 | 0.462 | 0.433 | 0.481 | 0.305 | 0.365 | 0.384 | 0.274 | 0.346 | 0.514 | 0.465 | 0.416 | 0.345 | 0.399 | 0.506 | 0.536 | 0.532 | 0.386 | 0.543 | 0.552 | 0.527 | 0.431 | 0.474 | 0.579 | 0.597 | 0.592 | 0.351 | 0.142 | 0.313 | 0.539 |
| Q4_5 | 0.574 | 0.731 | 0.550 | 0.590 | 1.000 | 0.581 | 0.357 | 0.590 | 0.474 | 0.576 | 0.602 | 0.576 | 0.635 | 0.521 | 0.618 | 0.516 | 0.570 | 0.685 | 0.610 | 0.507 | 0.444 | 0.492 | 0.412 | 0.318 | 0.440 | 0.421 | 0.466 | 0.600 | 0.461 | 0.558 | 0.465 | 0.400 | 0.426 | 0.440 | 0.329 | 0.322 | 0.536 | 0.501 | 0.451 | 0.338 | 0.419 | 0.518 | 0.578 | 0.446 | 0.318 | 0.420 | 0.474 | 0.424 | 0.419 | 0.424 | 0.559 | 0.621 | 0.409 | 0.315 | 0.078 | 0.172 | 0.543 |
| Q4_6 | 0.522 | 0.551 | 0.463 | 0.544 | 0.581 | 1.000 | 0.640 | 0.477 | 0.377 | 0.529 | 0.498 | 0.442 | 0.396 | 0.451 | 0.502 | 0.516 | 0.448 | 0.563 | 0.500 | 0.413 | 0.523 | 0.486 | 0.431 | 0.359 | 0.501 | 0.419 | 0.477 | 0.545 | 0.383 | 0.457 | 0.466 | 0.376 | 0.490 | 0.425 | 0.338 | 0.314 | 0.413 | 0.417 | 0.369 | 0.313 | 0.432 | 0.529 | 0.552 | 0.466 | 0.413 | 0.456 | 0.434 | 0.432 | 0.435 | 0.360 | 0.497 | 0.563 | 0.426 | 0.329 | 0.251 | 0.431 | 0.523 |
| Q4_7 | 0.349 | 0.380 | 0.347 | 0.469 | 0.357 | 0.640 | 1.000 | 0.334 | 0.289 | 0.442 | 0.460 | 0.349 | 0.176 | 0.397 | 0.319 | 0.399 | 0.325 | 0.351 | 0.405 | 0.373 | 0.423 | 0.341 | 0.431 | 0.326 | 0.385 | 0.434 | 0.448 | 0.405 | 0.385 | 0.377 | 0.431 | 0.253 | 0.321 | 0.284 | 0.300 | 0.186 | 0.360 | 0.338 | 0.343 | 0.279 | 0.376 | 0.473 | 0.389 | 0.464 | 0.482 | 0.439 | 0.444 | 0.383 | 0.461 | 0.358 | 0.369 | 0.363 | 0.363 | 0.278 | 0.217 | 0.337 | 0.332 |
| Q4_8 | 0.655 | 0.693 | 0.681 | 0.690 | 0.590 | 0.477 | 0.334 | 1.000 | 0.652 | 0.654 | 0.636 | 0.565 | 0.583 | 0.572 | 0.650 | 0.529 | 0.627 | 0.719 | 0.693 | 0.467 | 0.398 | 0.452 | 0.301 | 0.247 | 0.408 | 0.351 | 0.423 | 0.486 | 0.382 | 0.450 | 0.399 | 0.355 | 0.366 | 0.425 | 0.347 | 0.343 | 0.481 | 0.556 | 0.461 | 0.361 | 0.399 | 0.463 | 0.476 | 0.429 | 0.281 | 0.333 | 0.420 | 0.419 | 0.316 | 0.411 | 0.490 | 0.552 | 0.461 | 0.271 | 0.085 | 0.166 | 0.446 |
| Q4_9 | 0.491 | 0.576 | 0.579 | 0.572 | 0.474 | 0.377 | 0.289 | 0.652 | 1.000 | 0.540 | 0.594 | 0.519 | 0.548 | 0.502 | 0.600 | 0.475 | 0.543 | 0.613 | 0.766 | 0.438 | 0.390 | 0.413 | 0.267 | 0.284 | 0.414 | 0.329 | 0.359 | 0.382 | 0.316 | 0.451 | 0.450 | 0.328 | 0.336 | 0.324 | 0.319 | 0.305 | 0.394 | 0.474 | 0.384 | 0.375 | 0.308 | 0.350 | 0.439 | 0.451 | 0.332 | 0.370 | 0.470 | 0.506 | 0.392 | 0.422 | 0.467 | 0.519 | 0.534 | 0.360 | 0.135 | 0.213 | 0.384 |
| Q4_10 | 0.643 | 0.629 | 0.676 | 0.735 | 0.576 | 0.529 | 0.442 | 0.654 | 0.540 | 1.000 | 0.642 | 0.519 | 0.456 | 0.497 | 0.611 | 0.568 | 0.552 | 0.721 | 0.640 | 0.507 | 0.324 | 0.361 | 0.375 | 0.376 | 0.436 | 0.442 | 0.418 | 0.538 | 0.446 | 0.459 | 0.426 | 0.341 | 0.360 | 0.436 | 0.332 | 0.322 | 0.486 | 0.540 | 0.418 | 0.378 | 0.334 | 0.502 | 0.485 | 0.572 | 0.419 | 0.440 | 0.545 | 0.449 | 0.441 | 0.458 | 0.442 | 0.521 | 0.559 | 0.333 | 0.176 | 0.338 | 0.506 |
| Q4_11 | 0.648 | 0.632 | 0.632 | 0.642 | 0.602 | 0.498 | 0.460 | 0.636 | 0.594 | 0.642 | 1.000 | 0.634 | 0.588 | 0.600 | 0.653 | 0.619 | 0.595 | 0.724 | 0.738 | 0.522 | 0.441 | 0.454 | 0.353 | 0.302 | 0.473 | 0.491 | 0.457 | 0.585 | 0.550 | 0.512 | 0.437 | 0.400 | 0.397 | 0.396 | 0.370 | 0.414 | 0.500 | 0.520 | 0.510 | 0.437 | 0.413 | 0.504 | 0.589 | 0.511 | 0.367 | 0.435 | 0.599 | 0.512 | 0.399 | 0.459 | 0.485 | 0.539 | 0.544 | 0.432 | 0.226 | 0.312 | 0.566 |
| Q4_12 | 0.471 | 0.557 | 0.409 | 0.521 | 0.576 | 0.442 | 0.349 | 0.565 | 0.519 | 0.519 | 0.634 | 1.000 | 0.814 | 0.608 | 0.568 | 0.490 | 0.562 | 0.632 | 0.603 | 0.454 | 0.434 | 0.481 | 0.290 | 0.213 | 0.370 | 0.491 | 0.415 | 0.489 | 0.463 | 0.411 | 0.404 | 0.520 | 0.505 | 0.517 | 0.416 | 0.421 | 0.599 | 0.610 | 0.600 | 0.467 | 0.392 | 0.551 | 0.520 | 0.525 | 0.365 | 0.408 | 0.521 | 0.485 | 0.375 | 0.421 | 0.458 | 0.454 | 0.342 | 0.304 | 0.110 | 0.193 | 0.428 |
| Q4_13 | 0.500 | 0.615 | 0.451 | 0.467 | 0.635 | 0.396 | 0.176 | 0.583 | 0.548 | 0.456 | 0.588 | 0.814 | 1.000 | 0.611 | 0.577 | 0.458 | 0.521 | 0.698 | 0.619 | 0.514 | 0.446 | 0.503 | 0.236 | 0.179 | 0.361 | 0.476 | 0.356 | 0.496 | 0.463 | 0.452 | 0.427 | 0.561 | 0.506 | 0.490 | 0.427 | 0.463 | 0.575 | 0.574 | 0.521 | 0.445 | 0.315 | 0.452 | 0.529 | 0.485 | 0.226 | 0.288 | 0.385 | 0.471 | 0.258 | 0.359 | 0.488 | 0.558 | 0.352 | 0.345 | 0.062 | 0.083 | 0.398 |
| Q4_14 | 0.607 | 0.534 | 0.502 | 0.537 | 0.521 | 0.451 | 0.397 | 0.572 | 0.502 | 0.497 | 0.600 | 0.608 | 0.611 | 1.000 | 0.742 | 0.655 | 0.502 | 0.633 | 0.580 | 0.519 | 0.469 | 0.519 | 0.343 | 0.284 | 0.509 | 0.445 | 0.417 | 0.560 | 0.406 | 0.516 | 0.451 | 0.505 | 0.430 | 0.431 | 0.411 | 0.401 | 0.501 | 0.567 | 0.518 | 0.508 | 0.486 | 0.576 | 0.535 | 0.484 | 0.364 | 0.365 | 0.507 | 0.519 | 0.424 | 0.431 | 0.546 | 0.491 | 0.433 | 0.411 | 0.240 | 0.278 | 0.487 |
| Q4_15 | 0.602 | 0.663 | 0.603 | 0.589 | 0.618 | 0.502 | 0.319 | 0.650 | 0.600 | 0.611 | 0.653 | 0.568 | 0.577 | 0.742 | 1.000 | 0.774 | 0.671 | 0.735 | 0.659 | 0.526 | 0.451 | 0.514 | 0.401 | 0.320 | 0.551 | 0.500 | 0.478 | 0.593 | 0.505 | 0.517 | 0.504 | 0.421 | 0.408 | 0.416 | 0.384 | 0.364 | 0.508 | 0.557 | 0.507 | 0.432 | 0.435 | 0.503 | 0.566 | 0.521 | 0.294 | 0.373 | 0.532 | 0.519 | 0.384 | 0.484 | 0.503 | 0.553 | 0.557 | 0.370 | 0.145 | 0.207 | 0.501 |
| Q4_16 | 0.606 | 0.565 | 0.571 | 0.592 | 0.516 | 0.516 | 0.399 | 0.529 | 0.475 | 0.568 | 0.619 | 0.490 | 0.458 | 0.655 | 0.774 | 1.000 | 0.669 | 0.646 | 0.594 | 0.461 | 0.445 | 0.524 | 0.388 | 0.356 | 0.554 | 0.468 | 0.443 | 0.557 | 0.463 | 0.497 | 0.473 | 0.499 | 0.492 | 0.448 | 0.417 | 0.246 | 0.462 | 0.513 | 0.447 | 0.325 | 0.408 | 0.437 | 0.524 | 0.426 | 0.253 | 0.328 | 0.431 | 0.437 | 0.288 | 0.364 | 0.486 | 0.506 | 0.438 | 0.291 | 0.106 | 0.160 | 0.483 |
| Q4_17 | 0.533 | 0.591 | 0.542 | 0.591 | 0.570 | 0.448 | 0.325 | 0.627 | 0.543 | 0.552 | 0.595 | 0.562 | 0.521 | 0.502 | 0.671 | 0.669 | 1.000 | 0.691 | 0.601 | 0.440 | 0.381 | 0.421 | 0.267 | 0.190 | 0.421 | 0.418 | 0.355 | 0.404 | 0.469 | 0.451 | 0.382 | 0.394 | 0.350 | 0.427 | 0.354 | 0.313 | 0.498 | 0.525 | 0.469 | 0.344 | 0.436 | 0.408 | 0.431 | 0.397 | 0.318 | 0.407 | 0.490 | 0.420 | 0.382 | 0.450 | 0.491 | 0.512 | 0.470 | 0.301 | 0.090 | 0.160 | 0.455 |
| Q4_18 | 0.734 | 0.804 | 0.694 | 0.749 | 0.685 | 0.563 | 0.351 | 0.719 | 0.613 | 0.721 | 0.724 | 0.632 | 0.698 | 0.633 | 0.735 | 0.646 | 0.691 | 1.000 | 0.731 | 0.547 | 0.483 | 0.479 | 0.365 | 0.369 | 0.585 | 0.452 | 0.495 | 0.605 | 0.587 | 0.505 | 0.480 | 0.380 | 0.361 | 0.406 | 0.325 | 0.377 | 0.472 | 0.566 | 0.454 | 0.390 | 0.420 | 0.535 | 0.569 | 0.520 | 0.336 | 0.440 | 0.506 | 0.469 | 0.406 | 0.465 | 0.541 | 0.626 | 0.562 | 0.312 | 0.096 | 0.295 | 0.584 |
| Q4_19 | 0.558 | 0.624 | 0.640 | 0.594 | 0.610 | 0.500 | 0.405 | 0.693 | 0.766 | 0.640 | 0.738 | 0.603 | 0.619 | 0.580 | 0.659 | 0.594 | 0.601 | 0.731 | 1.000 | 0.528 | 0.501 | 0.522 | 0.357 | 0.307 | 0.530 | 0.461 | 0.468 | 0.586 | 0.462 | 0.522 | 0.494 | 0.417 | 0.404 | 0.416 | 0.391 | 0.328 | 0.505 | 0.541 | 0.466 | 0.449 | 0.456 | 0.505 | 0.561 | 0.507 | 0.422 | 0.431 | 0.553 | 0.537 | 0.490 | 0.542 | 0.547 | 0.569 | 0.553 | 0.419 | 0.134 | 0.266 | 0.515 |
| Q5_1 | 0.474 | 0.530 | 0.485 | 0.474 | 0.507 | 0.413 | 0.373 | 0.467 | 0.438 | 0.507 | 0.522 | 0.454 | 0.514 | 0.519 | 0.526 | 0.461 | 0.440 | 0.547 | 0.528 | 1.000 | 0.555 | 0.631 | 0.438 | 0.366 | 0.550 | 0.432 | 0.514 | 0.608 | 0.530 | 0.562 | 0.517 | 0.381 | 0.315 | 0.291 | 0.235 | 0.360 | 0.424 | 0.432 | 0.352 | 0.418 | 0.330 | 0.465 | 0.593 | 0.474 | 0.334 | 0.361 | 0.515 | 0.441 | 0.407 | 0.419 | 0.542 | 0.474 | 0.530 | 0.423 | 0.167 | 0.270 | 0.487 |
| Q5_2 | 0.377 | 0.434 | 0.387 | 0.401 | 0.444 | 0.523 | 0.423 | 0.398 | 0.390 | 0.324 | 0.441 | 0.434 | 0.446 | 0.469 | 0.451 | 0.445 | 0.381 | 0.483 | 0.501 | 0.555 | 1.000 | 0.707 | 0.503 | 0.447 | 0.659 | 0.443 | 0.578 | 0.515 | 0.485 | 0.411 | 0.544 | 0.379 | 0.391 | 0.383 | 0.404 | 0.311 | 0.396 | 0.403 | 0.377 | 0.300 | 0.377 | 0.533 | 0.511 | 0.484 | 0.384 | 0.419 | 0.432 | 0.459 | 0.484 | 0.341 | 0.475 | 0.474 | 0.359 | 0.258 | 0.164 | 0.327 | 0.444 |
| Q5_3 | 0.407 | 0.472 | 0.400 | 0.399 | 0.492 | 0.486 | 0.341 | 0.452 | 0.413 | 0.361 | 0.454 | 0.481 | 0.503 | 0.519 | 0.514 | 0.524 | 0.421 | 0.479 | 0.522 | 0.631 | 0.707 | 1.000 | 0.569 | 0.441 | 0.671 | 0.427 | 0.625 | 0.631 | 0.487 | 0.536 | 0.538 | 0.453 | 0.369 | 0.357 | 0.345 | 0.298 | 0.427 | 0.424 | 0.410 | 0.365 | 0.373 | 0.469 | 0.576 | 0.471 | 0.300 | 0.332 | 0.429 | 0.423 | 0.369 | 0.374 | 0.512 | 0.475 | 0.388 | 0.316 | 0.129 | 0.246 | 0.486 |
| Q5_4 | 0.381 | 0.384 | 0.357 | 0.414 | 0.412 | 0.431 | 0.431 | 0.301 | 0.267 | 0.375 | 0.353 | 0.290 | 0.236 | 0.343 | 0.401 | 0.388 | 0.267 | 0.365 | 0.357 | 0.438 | 0.503 | 0.569 | 1.000 | 0.837 | 0.632 | 0.390 | 0.585 | 0.488 | 0.433 | 0.417 | 0.486 | 0.221 | 0.267 | 0.290 | 0.205 | 0.243 | 0.302 | 0.227 | 0.210 | 0.223 | 0.320 | 0.415 | 0.451 | 0.444 | 0.478 | 0.324 | 0.389 | 0.380 | 0.532 | 0.364 | 0.371 | 0.367 | 0.373 | 0.341 | 0.168 | 0.379 | 0.344 |
| Q5_5 | 0.319 | 0.338 | 0.328 | 0.399 | 0.318 | 0.359 | 0.326 | 0.247 | 0.284 | 0.376 | 0.302 | 0.213 | 0.179 | 0.284 | 0.320 | 0.356 | 0.190 | 0.369 | 0.307 | 0.366 | 0.447 | 0.441 | 0.837 | 1.000 | 0.628 | 0.367 | 0.577 | 0.446 | 0.343 | 0.408 | 0.433 | 0.141 | 0.208 | 0.228 | 0.180 | 0.226 | 0.205 | 0.180 | 0.140 | 0.153 | 0.237 | 0.289 | 0.394 | 0.434 | 0.389 | 0.315 | 0.316 | 0.323 | 0.429 | 0.349 | 0.313 | 0.311 | 0.373 | 0.355 | 0.174 | 0.363 | 0.300 |
| Q5_6 | 0.438 | 0.479 | 0.488 | 0.523 | 0.440 | 0.501 | 0.385 | 0.408 | 0.414 | 0.436 | 0.473 | 0.370 | 0.361 | 0.509 | 0.551 | 0.554 | 0.421 | 0.585 | 0.530 | 0.550 | 0.659 | 0.671 | 0.632 | 0.628 | 1.000 | 0.561 | 0.717 | 0.632 | 0.488 | 0.487 | 0.570 | 0.305 | 0.373 | 0.400 | 0.331 | 0.352 | 0.386 | 0.464 | 0.408 | 0.289 | 0.442 | 0.495 | 0.538 | 0.463 | 0.423 | 0.435 | 0.510 | 0.488 | 0.470 | 0.426 | 0.506 | 0.519 | 0.481 | 0.374 | 0.150 | 0.376 | 0.452 |
| Q5_7 | 0.340 | 0.351 | 0.348 | 0.446 | 0.421 | 0.419 | 0.434 | 0.351 | 0.329 | 0.442 | 0.491 | 0.491 | 0.476 | 0.445 | 0.500 | 0.468 | 0.418 | 0.452 | 0.461 | 0.432 | 0.443 | 0.427 | 0.390 | 0.367 | 0.561 | 1.000 | 0.572 | 0.627 | 0.576 | 0.460 | 0.475 | 0.429 | 0.499 | 0.520 | 0.396 | 0.374 | 0.514 | 0.590 | 0.533 | 0.467 | 0.399 | 0.541 | 0.520 | 0.546 | 0.384 | 0.394 | 0.468 | 0.453 | 0.341 | 0.457 | 0.419 | 0.420 | 0.452 | 0.325 | 0.151 | 0.215 | 0.369 |
| Q5_8 | 0.347 | 0.423 | 0.388 | 0.422 | 0.466 | 0.477 | 0.448 | 0.423 | 0.359 | 0.418 | 0.457 | 0.415 | 0.356 | 0.417 | 0.478 | 0.443 | 0.355 | 0.495 | 0.468 | 0.514 | 0.578 | 0.625 | 0.585 | 0.577 | 0.717 | 0.572 | 1.000 | 0.693 | 0.532 | 0.520 | 0.534 | 0.325 | 0.378 | 0.429 | 0.321 | 0.411 | 0.392 | 0.399 | 0.399 | 0.291 | 0.421 | 0.534 | 0.553 | 0.496 | 0.440 | 0.460 | 0.465 | 0.488 | 0.426 | 0.464 | 0.473 | 0.472 | 0.460 | 0.385 | 0.169 | 0.344 | 0.422 |
| Q5_9 | 0.487 | 0.512 | 0.505 | 0.485 | 0.600 | 0.545 | 0.405 | 0.486 | 0.382 | 0.538 | 0.585 | 0.489 | 0.496 | 0.560 | 0.593 | 0.557 | 0.404 | 0.605 | 0.586 | 0.608 | 0.515 | 0.631 | 0.488 | 0.446 | 0.632 | 0.627 | 0.693 | 1.000 | 0.627 | 0.599 | 0.568 | 0.498 | 0.494 | 0.511 | 0.390 | 0.389 | 0.519 | 0.550 | 0.510 | 0.421 | 0.502 | 0.620 | 0.645 | 0.512 | 0.379 | 0.478 | 0.498 | 0.444 | 0.344 | 0.463 | 0.577 | 0.557 | 0.496 | 0.376 | 0.103 | 0.278 | 0.565 |
| Q5_10 | 0.404 | 0.455 | 0.385 | 0.462 | 0.461 | 0.383 | 0.385 | 0.382 | 0.316 | 0.446 | 0.550 | 0.463 | 0.463 | 0.406 | 0.505 | 0.463 | 0.469 | 0.587 | 0.462 | 0.530 | 0.485 | 0.487 | 0.433 | 0.343 | 0.488 | 0.576 | 0.532 | 0.627 | 1.000 | 0.498 | 0.470 | 0.345 | 0.337 | 0.423 | 0.351 | 0.283 | 0.525 | 0.462 | 0.458 | 0.328 | 0.398 | 0.464 | 0.500 | 0.476 | 0.295 | 0.405 | 0.473 | 0.446 | 0.386 | 0.459 | 0.454 | 0.498 | 0.478 | 0.322 | 0.039 | 0.284 | 0.397 |
| Q5_11 | 0.476 | 0.532 | 0.538 | 0.433 | 0.558 | 0.457 | 0.377 | 0.450 | 0.451 | 0.459 | 0.512 | 0.411 | 0.452 | 0.516 | 0.517 | 0.497 | 0.451 | 0.505 | 0.522 | 0.562 | 0.411 | 0.536 | 0.417 | 0.408 | 0.487 | 0.460 | 0.520 | 0.599 | 0.498 | 1.000 | 0.549 | 0.412 | 0.428 | 0.384 | 0.332 | 0.425 | 0.440 | 0.461 | 0.422 | 0.380 | 0.478 | 0.461 | 0.637 | 0.479 | 0.432 | 0.491 | 0.501 | 0.442 | 0.423 | 0.419 | 0.581 | 0.528 | 0.507 | 0.510 | 0.353 | 0.309 | 0.473 |
| Q5_12 | 0.364 | 0.421 | 0.323 | 0.481 | 0.465 | 0.466 | 0.431 | 0.399 | 0.450 | 0.426 | 0.437 | 0.404 | 0.427 | 0.451 | 0.504 | 0.473 | 0.382 | 0.480 | 0.494 | 0.517 | 0.544 | 0.538 | 0.486 | 0.433 | 0.570 | 0.475 | 0.534 | 0.568 | 0.470 | 0.549 | 1.000 | 0.343 | 0.483 | 0.411 | 0.324 | 0.331 | 0.453 | 0.460 | 0.402 | 0.314 | 0.400 | 0.521 | 0.536 | 0.429 | 0.430 | 0.520 | 0.515 | 0.423 | 0.441 | 0.485 | 0.542 | 0.521 | 0.475 | 0.414 | 0.217 | 0.349 | 0.494 |
| Q6_1 | 0.370 | 0.351 | 0.393 | 0.305 | 0.400 | 0.376 | 0.253 | 0.355 | 0.328 | 0.341 | 0.400 | 0.520 | 0.561 | 0.505 | 0.421 | 0.499 | 0.394 | 0.380 | 0.417 | 0.381 | 0.379 | 0.453 | 0.221 | 0.141 | 0.305 | 0.429 | 0.325 | 0.498 | 0.345 | 0.412 | 0.343 | 1.000 | 0.788 | 0.753 | 0.663 | 0.380 | 0.675 | 0.671 | 0.666 | 0.352 | 0.351 | 0.444 | 0.572 | 0.384 | 0.142 | 0.277 | 0.344 | 0.366 | 0.166 | 0.231 | 0.478 | 0.470 | 0.308 | 0.319 | 0.134 | 0.071 | 0.464 |
| Q6_2 | 0.325 | 0.372 | 0.313 | 0.365 | 0.426 | 0.490 | 0.321 | 0.366 | 0.336 | 0.360 | 0.397 | 0.505 | 0.506 | 0.430 | 0.408 | 0.492 | 0.350 | 0.361 | 0.404 | 0.315 | 0.391 | 0.369 | 0.267 | 0.208 | 0.373 | 0.499 | 0.378 | 0.494 | 0.337 | 0.428 | 0.483 | 0.788 | 1.000 | 0.803 | 0.787 | 0.429 | 0.671 | 0.669 | 0.707 | 0.298 | 0.354 | 0.476 | 0.523 | 0.405 | 0.261 | 0.361 | 0.392 | 0.399 | 0.159 | 0.252 | 0.454 | 0.486 | 0.333 | 0.256 | 0.081 | 0.145 | 0.388 |
| Q6_3 | 0.304 | 0.337 | 0.317 | 0.384 | 0.440 | 0.425 | 0.284 | 0.425 | 0.324 | 0.436 | 0.396 | 0.517 | 0.490 | 0.431 | 0.416 | 0.448 | 0.427 | 0.406 | 0.416 | 0.291 | 0.383 | 0.357 | 0.290 | 0.228 | 0.400 | 0.520 | 0.429 | 0.511 | 0.423 | 0.384 | 0.411 | 0.753 | 0.803 | 1.000 | 0.743 | 0.459 | 0.737 | 0.767 | 0.664 | 0.295 | 0.316 | 0.492 | 0.535 | 0.391 | 0.319 | 0.349 | 0.361 | 0.408 | 0.212 | 0.255 | 0.412 | 0.481 | 0.318 | 0.230 | 0.073 | 0.163 | 0.415 |
| Q6_4 | 0.307 | 0.285 | 0.284 | 0.274 | 0.329 | 0.338 | 0.300 | 0.347 | 0.319 | 0.332 | 0.370 | 0.416 | 0.427 | 0.411 | 0.384 | 0.417 | 0.354 | 0.325 | 0.391 | 0.235 | 0.404 | 0.345 | 0.205 | 0.180 | 0.331 | 0.396 | 0.321 | 0.390 | 0.351 | 0.332 | 0.324 | 0.663 | 0.787 | 0.743 | 1.000 | 0.310 | 0.638 | 0.664 | 0.719 | 0.226 | 0.338 | 0.422 | 0.403 | 0.361 | 0.208 | 0.360 | 0.301 | 0.368 | 0.140 | 0.239 | 0.373 | 0.448 | 0.349 | 0.165 | 0.003 | 0.075 | 0.311 |
| Q6_5 | 0.300 | 0.400 | 0.360 | 0.346 | 0.322 | 0.314 | 0.186 | 0.343 | 0.305 | 0.322 | 0.414 | 0.421 | 0.463 | 0.401 | 0.364 | 0.246 | 0.313 | 0.377 | 0.328 | 0.360 | 0.311 | 0.298 | 0.243 | 0.226 | 0.352 | 0.374 | 0.411 | 0.389 | 0.283 | 0.425 | 0.331 | 0.380 | 0.429 | 0.459 | 0.310 | 1.000 | 0.486 | 0.498 | 0.466 | 0.386 | 0.477 | 0.404 | 0.455 | 0.428 | 0.237 | 0.291 | 0.341 | 0.351 | 0.262 | 0.312 | 0.436 | 0.472 | 0.396 | 0.357 | 0.213 | 0.169 | 0.408 |
| Q6_6 | 0.356 | 0.475 | 0.363 | 0.514 | 0.536 | 0.413 | 0.360 | 0.481 | 0.394 | 0.486 | 0.500 | 0.599 | 0.575 | 0.501 | 0.508 | 0.462 | 0.498 | 0.472 | 0.505 | 0.424 | 0.396 | 0.427 | 0.302 | 0.205 | 0.386 | 0.514 | 0.392 | 0.519 | 0.525 | 0.440 | 0.453 | 0.675 | 0.671 | 0.737 | 0.638 | 0.486 | 1.000 | 0.786 | 0.778 | 0.429 | 0.452 | 0.641 | 0.609 | 0.526 | 0.280 | 0.465 | 0.486 | 0.464 | 0.226 | 0.447 | 0.518 | 0.613 | 0.502 | 0.260 | 0.041 | 0.078 | 0.536 |
| Q6_7 | 0.434 | 0.468 | 0.432 | 0.465 | 0.501 | 0.417 | 0.338 | 0.556 | 0.474 | 0.540 | 0.520 | 0.610 | 0.574 | 0.567 | 0.557 | 0.513 | 0.525 | 0.566 | 0.541 | 0.432 | 0.403 | 0.424 | 0.227 | 0.180 | 0.464 | 0.590 | 0.399 | 0.550 | 0.462 | 0.461 | 0.460 | 0.671 | 0.669 | 0.767 | 0.664 | 0.498 | 0.786 | 1.000 | 0.814 | 0.468 | 0.501 | 0.640 | 0.589 | 0.520 | 0.347 | 0.439 | 0.502 | 0.461 | 0.311 | 0.441 | 0.506 | 0.572 | 0.485 | 0.298 | 0.062 | 0.141 | 0.515 |
| Q6_8 | 0.370 | 0.359 | 0.357 | 0.416 | 0.451 | 0.369 | 0.343 | 0.461 | 0.384 | 0.418 | 0.510 | 0.600 | 0.521 | 0.518 | 0.507 | 0.447 | 0.469 | 0.454 | 0.466 | 0.352 | 0.377 | 0.410 | 0.210 | 0.140 | 0.408 | 0.533 | 0.399 | 0.510 | 0.458 | 0.422 | 0.402 | 0.666 | 0.707 | 0.664 | 0.719 | 0.466 | 0.778 | 0.814 | 1.000 | 0.469 | 0.465 | 0.568 | 0.544 | 0.469 | 0.278 | 0.488 | 0.487 | 0.482 | 0.215 | 0.387 | 0.498 | 0.538 | 0.477 | 0.274 | 0.048 | 0.110 | 0.516 |
| Q6_9 | 0.288 | 0.318 | 0.290 | 0.345 | 0.338 | 0.313 | 0.279 | 0.361 | 0.375 | 0.378 | 0.437 | 0.467 | 0.445 | 0.508 | 0.432 | 0.325 | 0.344 | 0.390 | 0.449 | 0.418 | 0.300 | 0.365 | 0.223 | 0.153 | 0.289 | 0.467 | 0.291 | 0.421 | 0.328 | 0.380 | 0.314 | 0.352 | 0.298 | 0.295 | 0.226 | 0.386 | 0.429 | 0.468 | 0.469 | 1.000 | 0.539 | 0.534 | 0.454 | 0.422 | 0.394 | 0.323 | 0.496 | 0.507 | 0.402 | 0.416 | 0.394 | 0.402 | 0.371 | 0.430 | 0.389 | 0.275 | 0.400 |
| Q6_10 | 0.345 | 0.410 | 0.319 | 0.399 | 0.419 | 0.432 | 0.376 | 0.399 | 0.308 | 0.334 | 0.413 | 0.392 | 0.315 | 0.486 | 0.435 | 0.408 | 0.436 | 0.420 | 0.456 | 0.330 | 0.377 | 0.373 | 0.320 | 0.237 | 0.442 | 0.399 | 0.421 | 0.502 | 0.398 | 0.478 | 0.400 | 0.351 | 0.354 | 0.316 | 0.338 | 0.477 | 0.452 | 0.501 | 0.465 | 0.539 | 1.000 | 0.630 | 0.482 | 0.393 | 0.323 | 0.350 | 0.456 | 0.433 | 0.429 | 0.427 | 0.505 | 0.481 | 0.442 | 0.437 | 0.231 | 0.284 | 0.485 |
| Q6_11 | 0.404 | 0.483 | 0.415 | 0.506 | 0.518 | 0.529 | 0.473 | 0.463 | 0.350 | 0.502 | 0.504 | 0.551 | 0.452 | 0.576 | 0.503 | 0.437 | 0.408 | 0.535 | 0.505 | 0.465 | 0.533 | 0.469 | 0.415 | 0.289 | 0.495 | 0.541 | 0.534 | 0.620 | 0.464 | 0.461 | 0.521 | 0.444 | 0.476 | 0.492 | 0.422 | 0.404 | 0.641 | 0.640 | 0.568 | 0.534 | 0.630 | 1.000 | 0.627 | 0.543 | 0.510 | 0.501 | 0.571 | 0.532 | 0.508 | 0.469 | 0.533 | 0.539 | 0.444 | 0.392 | 0.240 | 0.399 | 0.525 |
| Q7_1 | 0.516 | 0.557 | 0.504 | 0.536 | 0.578 | 0.552 | 0.389 | 0.476 | 0.439 | 0.485 | 0.589 | 0.520 | 0.529 | 0.535 | 0.566 | 0.524 | 0.431 | 0.569 | 0.561 | 0.593 | 0.511 | 0.576 | 0.451 | 0.394 | 0.538 | 0.520 | 0.553 | 0.645 | 0.500 | 0.637 | 0.536 | 0.572 | 0.523 | 0.535 | 0.403 | 0.455 | 0.609 | 0.589 | 0.544 | 0.454 | 0.482 | 0.627 | 1.000 | 0.706 | 0.489 | 0.582 | 0.612 | 0.575 | 0.445 | 0.502 | 0.689 | 0.722 | 0.602 | 0.488 | 0.214 | 0.273 | 0.702 |
| Q7_2 | 0.390 | 0.453 | 0.398 | 0.532 | 0.446 | 0.466 | 0.464 | 0.429 | 0.451 | 0.572 | 0.511 | 0.525 | 0.485 | 0.484 | 0.521 | 0.426 | 0.397 | 0.520 | 0.507 | 0.474 | 0.484 | 0.471 | 0.444 | 0.434 | 0.463 | 0.546 | 0.496 | 0.512 | 0.476 | 0.479 | 0.429 | 0.384 | 0.405 | 0.391 | 0.361 | 0.428 | 0.526 | 0.520 | 0.469 | 0.422 | 0.393 | 0.543 | 0.706 | 1.000 | 0.561 | 0.617 | 0.599 | 0.546 | 0.508 | 0.544 | 0.530 | 0.568 | 0.649 | 0.408 | 0.247 | 0.378 | 0.570 |
| Q7_3 | 0.266 | 0.315 | 0.308 | 0.386 | 0.318 | 0.413 | 0.482 | 0.281 | 0.332 | 0.419 | 0.367 | 0.365 | 0.226 | 0.364 | 0.294 | 0.253 | 0.318 | 0.336 | 0.422 | 0.334 | 0.384 | 0.300 | 0.478 | 0.389 | 0.423 | 0.384 | 0.440 | 0.379 | 0.295 | 0.432 | 0.430 | 0.142 | 0.261 | 0.319 | 0.208 | 0.237 | 0.280 | 0.347 | 0.278 | 0.394 | 0.323 | 0.510 | 0.489 | 0.561 | 1.000 | 0.609 | 0.569 | 0.589 | 0.803 | 0.498 | 0.325 | 0.328 | 0.430 | 0.498 | 0.351 | 0.497 | 0.370 |
| Q7_4 | 0.369 | 0.388 | 0.338 | 0.543 | 0.420 | 0.456 | 0.439 | 0.333 | 0.370 | 0.440 | 0.435 | 0.408 | 0.288 | 0.365 | 0.373 | 0.328 | 0.407 | 0.440 | 0.431 | 0.361 | 0.419 | 0.332 | 0.324 | 0.315 | 0.435 | 0.394 | 0.460 | 0.478 | 0.405 | 0.491 | 0.520 | 0.277 | 0.361 | 0.349 | 0.360 | 0.291 | 0.465 | 0.439 | 0.488 | 0.323 | 0.350 | 0.501 | 0.582 | 0.617 | 0.609 | 1.000 | 0.707 | 0.560 | 0.483 | 0.551 | 0.554 | 0.527 | 0.564 | 0.402 | 0.171 | 0.382 | 0.558 |
| Q7_5 | 0.444 | 0.546 | 0.454 | 0.552 | 0.474 | 0.434 | 0.444 | 0.420 | 0.470 | 0.545 | 0.599 | 0.521 | 0.385 | 0.507 | 0.532 | 0.431 | 0.490 | 0.506 | 0.553 | 0.515 | 0.432 | 0.429 | 0.389 | 0.316 | 0.510 | 0.468 | 0.465 | 0.498 | 0.473 | 0.501 | 0.515 | 0.344 | 0.392 | 0.361 | 0.301 | 0.341 | 0.486 | 0.502 | 0.487 | 0.496 | 0.456 | 0.571 | 0.612 | 0.599 | 0.569 | 0.707 | 1.000 | 0.641 | 0.635 | 0.657 | 0.604 | 0.620 | 0.628 | 0.504 | 0.342 | 0.437 | 0.562 |
| Q7_6 | 0.406 | 0.442 | 0.443 | 0.527 | 0.424 | 0.432 | 0.383 | 0.419 | 0.506 | 0.449 | 0.512 | 0.485 | 0.471 | 0.519 | 0.519 | 0.437 | 0.420 | 0.469 | 0.537 | 0.441 | 0.459 | 0.423 | 0.380 | 0.323 | 0.488 | 0.453 | 0.488 | 0.444 | 0.446 | 0.442 | 0.423 | 0.366 | 0.399 | 0.408 | 0.368 | 0.351 | 0.464 | 0.461 | 0.482 | 0.507 | 0.433 | 0.532 | 0.575 | 0.546 | 0.589 | 0.560 | 0.641 | 1.000 | 0.620 | 0.587 | 0.510 | 0.546 | 0.558 | 0.518 | 0.267 | 0.527 | 0.413 |
| Q7_7 | 0.334 | 0.411 | 0.411 | 0.431 | 0.419 | 0.435 | 0.461 | 0.316 | 0.392 | 0.441 | 0.399 | 0.375 | 0.258 | 0.424 | 0.384 | 0.288 | 0.382 | 0.406 | 0.490 | 0.407 | 0.484 | 0.369 | 0.532 | 0.429 | 0.470 | 0.341 | 0.426 | 0.344 | 0.386 | 0.423 | 0.441 | 0.166 | 0.159 | 0.212 | 0.140 | 0.262 | 0.226 | 0.311 | 0.215 | 0.402 | 0.429 | 0.508 | 0.445 | 0.508 | 0.803 | 0.483 | 0.635 | 0.620 | 1.000 | 0.639 | 0.428 | 0.381 | 0.493 | 0.575 | 0.446 | 0.588 | 0.333 |
| Q7_8 | 0.332 | 0.449 | 0.379 | 0.474 | 0.424 | 0.360 | 0.358 | 0.411 | 0.422 | 0.458 | 0.459 | 0.421 | 0.359 | 0.431 | 0.484 | 0.364 | 0.450 | 0.465 | 0.542 | 0.419 | 0.341 | 0.374 | 0.364 | 0.349 | 0.426 | 0.457 | 0.464 | 0.463 | 0.459 | 0.419 | 0.485 | 0.231 | 0.252 | 0.255 | 0.239 | 0.312 | 0.447 | 0.441 | 0.387 | 0.416 | 0.427 | 0.469 | 0.502 | 0.544 | 0.498 | 0.551 | 0.657 | 0.587 | 0.639 | 1.000 | 0.572 | 0.532 | 0.680 | 0.439 | 0.293 | 0.419 | 0.435 |
| Q7_9 | 0.586 | 0.537 | 0.546 | 0.579 | 0.559 | 0.497 | 0.369 | 0.490 | 0.467 | 0.442 | 0.485 | 0.458 | 0.488 | 0.546 | 0.503 | 0.486 | 0.491 | 0.541 | 0.547 | 0.542 | 0.475 | 0.512 | 0.371 | 0.313 | 0.506 | 0.419 | 0.473 | 0.577 | 0.454 | 0.581 | 0.542 | 0.478 | 0.454 | 0.412 | 0.373 | 0.436 | 0.518 | 0.506 | 0.498 | 0.394 | 0.505 | 0.533 | 0.689 | 0.530 | 0.325 | 0.554 | 0.604 | 0.510 | 0.428 | 0.572 | 1.000 | 0.762 | 0.632 | 0.405 | 0.204 | 0.296 | 0.654 |
| Q7_10 | 0.570 | 0.591 | 0.549 | 0.597 | 0.621 | 0.563 | 0.363 | 0.552 | 0.519 | 0.521 | 0.539 | 0.454 | 0.558 | 0.491 | 0.553 | 0.506 | 0.512 | 0.626 | 0.569 | 0.474 | 0.474 | 0.475 | 0.367 | 0.311 | 0.519 | 0.420 | 0.472 | 0.557 | 0.498 | 0.528 | 0.521 | 0.470 | 0.486 | 0.481 | 0.448 | 0.472 | 0.613 | 0.572 | 0.538 | 0.402 | 0.481 | 0.539 | 0.722 | 0.568 | 0.328 | 0.527 | 0.620 | 0.546 | 0.381 | 0.532 | 0.762 | 1.000 | 0.624 | 0.358 | 0.132 | 0.213 | 0.761 |
| Q7_11 | 0.484 | 0.482 | 0.535 | 0.592 | 0.409 | 0.426 | 0.363 | 0.461 | 0.534 | 0.559 | 0.544 | 0.342 | 0.352 | 0.433 | 0.557 | 0.438 | 0.470 | 0.562 | 0.553 | 0.530 | 0.359 | 0.388 | 0.373 | 0.373 | 0.481 | 0.452 | 0.460 | 0.496 | 0.478 | 0.507 | 0.475 | 0.308 | 0.333 | 0.318 | 0.349 | 0.396 | 0.502 | 0.485 | 0.477 | 0.371 | 0.442 | 0.444 | 0.602 | 0.649 | 0.430 | 0.564 | 0.628 | 0.558 | 0.493 | 0.680 | 0.632 | 0.624 | 1.000 | 0.570 | 0.294 | 0.387 | 0.524 |
| Q7_12 | 0.351 | 0.314 | 0.374 | 0.351 | 0.315 | 0.329 | 0.278 | 0.271 | 0.360 | 0.333 | 0.432 | 0.304 | 0.345 | 0.411 | 0.370 | 0.291 | 0.301 | 0.312 | 0.419 | 0.423 | 0.258 | 0.316 | 0.341 | 0.355 | 0.374 | 0.325 | 0.385 | 0.376 | 0.322 | 0.510 | 0.414 | 0.319 | 0.256 | 0.230 | 0.165 | 0.357 | 0.260 | 0.298 | 0.274 | 0.430 | 0.437 | 0.392 | 0.488 | 0.408 | 0.498 | 0.402 | 0.504 | 0.518 | 0.575 | 0.439 | 0.405 | 0.358 | 0.570 | 1.000 | 0.559 | 0.492 | 0.397 |
| Q7_13 | 0.134 | 0.137 | 0.154 | 0.142 | 0.078 | 0.251 | 0.217 | 0.085 | 0.135 | 0.176 | 0.226 | 0.110 | 0.062 | 0.240 | 0.145 | 0.106 | 0.090 | 0.096 | 0.134 | 0.167 | 0.164 | 0.129 | 0.168 | 0.174 | 0.150 | 0.151 | 0.169 | 0.103 | 0.039 | 0.353 | 0.217 | 0.134 | 0.081 | 0.073 | 0.003 | 0.213 | 0.041 | 0.062 | 0.048 | 0.389 | 0.231 | 0.240 | 0.214 | 0.247 | 0.351 | 0.171 | 0.342 | 0.267 | 0.446 | 0.293 | 0.204 | 0.132 | 0.294 | 0.559 | 1.000 | 0.479 | 0.152 |
| Q7_14 | 0.229 | 0.217 | 0.230 | 0.313 | 0.172 | 0.431 | 0.337 | 0.166 | 0.213 | 0.338 | 0.312 | 0.193 | 0.083 | 0.278 | 0.207 | 0.160 | 0.160 | 0.295 | 0.266 | 0.270 | 0.327 | 0.246 | 0.379 | 0.363 | 0.376 | 0.215 | 0.344 | 0.278 | 0.284 | 0.309 | 0.349 | 0.071 | 0.145 | 0.163 | 0.075 | 0.169 | 0.078 | 0.141 | 0.110 | 0.275 | 0.284 | 0.399 | 0.273 | 0.378 | 0.497 | 0.382 | 0.437 | 0.527 | 0.588 | 0.419 | 0.296 | 0.213 | 0.387 | 0.492 | 0.479 | 1.000 | 0.297 |
| Q7_15 | 0.502 | 0.522 | 0.463 | 0.539 | 0.543 | 0.523 | 0.332 | 0.446 | 0.384 | 0.506 | 0.566 | 0.428 | 0.398 | 0.487 | 0.501 | 0.483 | 0.455 | 0.584 | 0.515 | 0.487 | 0.444 | 0.486 | 0.344 | 0.300 | 0.452 | 0.369 | 0.422 | 0.565 | 0.397 | 0.473 | 0.494 | 0.464 | 0.388 | 0.415 | 0.311 | 0.408 | 0.536 | 0.515 | 0.516 | 0.400 | 0.485 | 0.525 | 0.702 | 0.570 | 0.370 | 0.558 | 0.562 | 0.413 | 0.333 | 0.435 | 0.654 | 0.761 | 0.524 | 0.397 | 0.152 | 0.297 | 1.000 |
# Difference between correlation matrices
kable(corr.pc$rho-corr.ppm,digits=3,format = "html")%>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="500px")| Q4_1 | Q4_2 | Q4_3 | Q4_4 | Q4_5 | Q4_6 | Q4_7 | Q4_8 | Q4_9 | Q4_10 | Q4_11 | Q4_12 | Q4_13 | Q4_14 | Q4_15 | Q4_16 | Q4_17 | Q4_18 | Q4_19 | Q5_1 | Q5_2 | Q5_3 | Q5_4 | Q5_5 | Q5_6 | Q5_7 | Q5_8 | Q5_9 | Q5_10 | Q5_11 | Q5_12 | Q6_1 | Q6_2 | Q6_3 | Q6_4 | Q6_5 | Q6_6 | Q6_7 | Q6_8 | Q6_9 | Q6_10 | Q6_11 | Q7_1 | Q7_2 | Q7_3 | Q7_4 | Q7_5 | Q7_6 | Q7_7 | Q7_8 | Q7_9 | Q7_10 | Q7_11 | Q7_12 | Q7_13 | Q7_14 | Q7_15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q4_1 | 0.000 | 0.072 | 0.076 | 0.080 | 0.066 | 0.049 | 0.032 | 0.070 | 0.060 | 0.075 | 0.062 | 0.063 | 0.092 | 0.064 | 0.069 | 0.066 | 0.065 | 0.079 | 0.063 | 0.052 | 0.040 | 0.045 | 0.031 | 0.036 | 0.044 | 0.042 | 0.028 | 0.055 | 0.054 | 0.052 | 0.034 | 0.077 | 0.050 | 0.069 | 0.050 | 0.050 | 0.066 | 0.065 | 0.057 | 0.042 | 0.036 | 0.047 | 0.058 | 0.050 | 0.019 | 0.038 | 0.046 | 0.049 | 0.047 | 0.034 | 0.058 | 0.075 | 0.061 | 0.044 | 0.007 | 0.011 | 0.057 |
| Q4_2 | 0.072 | 0.000 | 0.076 | 0.081 | 0.073 | 0.065 | 0.039 | 0.072 | 0.062 | 0.077 | 0.069 | 0.069 | 0.097 | 0.050 | 0.071 | 0.064 | 0.067 | 0.074 | 0.063 | 0.066 | 0.053 | 0.059 | 0.049 | 0.046 | 0.065 | 0.043 | 0.054 | 0.066 | 0.076 | 0.060 | 0.053 | 0.086 | 0.057 | 0.066 | 0.042 | 0.060 | 0.084 | 0.076 | 0.058 | 0.052 | 0.046 | 0.061 | 0.074 | 0.066 | 0.030 | 0.046 | 0.072 | 0.058 | 0.059 | 0.055 | 0.066 | 0.086 | 0.075 | 0.034 | 0.008 | 0.020 | 0.069 |
| Q4_3 | 0.076 | 0.076 | 0.000 | 0.086 | 0.060 | 0.049 | 0.020 | 0.074 | 0.071 | 0.079 | 0.060 | 0.058 | 0.082 | 0.043 | 0.063 | 0.059 | 0.066 | 0.072 | 0.063 | 0.054 | 0.040 | 0.044 | 0.026 | 0.035 | 0.059 | 0.040 | 0.038 | 0.056 | 0.050 | 0.058 | 0.030 | 0.090 | 0.043 | 0.069 | 0.041 | 0.065 | 0.061 | 0.065 | 0.047 | 0.045 | 0.035 | 0.046 | 0.055 | 0.042 | 0.018 | 0.036 | 0.048 | 0.053 | 0.034 | 0.042 | 0.058 | 0.069 | 0.064 | 0.029 | 0.006 | 0.019 | 0.052 |
| Q4_4 | 0.080 | 0.081 | 0.086 | 0.000 | 0.072 | 0.055 | 0.039 | 0.081 | 0.074 | 0.090 | 0.068 | 0.083 | 0.085 | 0.048 | 0.064 | 0.066 | 0.084 | 0.086 | 0.064 | 0.056 | 0.033 | 0.051 | 0.031 | 0.034 | 0.049 | 0.040 | 0.035 | 0.056 | 0.052 | 0.049 | 0.044 | 0.071 | 0.059 | 0.085 | 0.047 | 0.065 | 0.106 | 0.071 | 0.062 | 0.055 | 0.035 | 0.045 | 0.067 | 0.057 | 0.016 | 0.056 | 0.053 | 0.054 | 0.028 | 0.038 | 0.058 | 0.081 | 0.075 | 0.017 | 0.008 | 0.006 | 0.062 |
| Q4_5 | 0.066 | 0.073 | 0.060 | 0.072 | 0.000 | 0.059 | 0.039 | 0.058 | 0.045 | 0.075 | 0.062 | 0.072 | 0.109 | 0.060 | 0.066 | 0.062 | 0.065 | 0.068 | 0.065 | 0.057 | 0.052 | 0.052 | 0.059 | 0.048 | 0.054 | 0.048 | 0.054 | 0.066 | 0.072 | 0.058 | 0.058 | 0.073 | 0.057 | 0.074 | 0.052 | 0.052 | 0.080 | 0.076 | 0.062 | 0.050 | 0.051 | 0.058 | 0.068 | 0.060 | 0.038 | 0.053 | 0.055 | 0.057 | 0.051 | 0.053 | 0.068 | 0.071 | 0.061 | 0.033 | -0.007 | -0.001 | 0.069 |
| Q4_6 | 0.049 | 0.065 | 0.049 | 0.055 | 0.059 | 0.000 | 0.070 | 0.056 | 0.042 | 0.061 | 0.047 | 0.053 | 0.079 | 0.045 | 0.050 | 0.054 | 0.052 | 0.066 | 0.054 | 0.040 | 0.044 | 0.046 | 0.037 | 0.033 | 0.044 | 0.040 | 0.036 | 0.052 | 0.036 | 0.042 | 0.040 | 0.060 | 0.055 | 0.060 | 0.038 | 0.034 | 0.063 | 0.054 | 0.045 | 0.037 | 0.045 | 0.043 | 0.056 | 0.051 | 0.032 | 0.038 | 0.034 | 0.038 | 0.065 | 0.033 | 0.049 | 0.065 | 0.050 | 0.029 | 0.026 | 0.031 | 0.057 |
| Q4_7 | 0.032 | 0.039 | 0.020 | 0.039 | 0.039 | 0.070 | 0.000 | 0.024 | 0.019 | 0.030 | 0.045 | 0.049 | 0.026 | 0.047 | 0.033 | 0.046 | 0.041 | 0.033 | 0.037 | 0.036 | 0.024 | 0.030 | 0.020 | 0.009 | 0.043 | 0.065 | 0.034 | 0.049 | 0.042 | 0.041 | 0.035 | 0.048 | 0.050 | 0.053 | 0.045 | 0.024 | 0.065 | 0.043 | 0.057 | 0.040 | 0.039 | 0.045 | 0.047 | 0.050 | 0.030 | 0.039 | 0.035 | 0.025 | 0.035 | 0.018 | 0.035 | 0.051 | 0.041 | 0.012 | 0.018 | 0.002 | 0.048 |
| Q4_8 | 0.070 | 0.072 | 0.074 | 0.081 | 0.058 | 0.056 | 0.024 | 0.000 | 0.057 | 0.081 | 0.062 | 0.067 | 0.097 | 0.060 | 0.064 | 0.059 | 0.068 | 0.077 | 0.073 | 0.050 | 0.047 | 0.052 | 0.030 | 0.030 | 0.048 | 0.043 | 0.048 | 0.053 | 0.057 | 0.046 | 0.045 | 0.069 | 0.046 | 0.073 | 0.045 | 0.052 | 0.071 | 0.067 | 0.054 | 0.051 | 0.046 | 0.057 | 0.061 | 0.059 | 0.028 | 0.037 | 0.049 | 0.055 | 0.047 | 0.052 | 0.056 | 0.064 | 0.064 | 0.025 | 0.003 | 0.010 | 0.049 |
| Q4_9 | 0.060 | 0.062 | 0.071 | 0.074 | 0.045 | 0.042 | 0.019 | 0.057 | 0.000 | 0.073 | 0.053 | 0.053 | 0.092 | 0.051 | 0.064 | 0.054 | 0.055 | 0.066 | 0.060 | 0.051 | 0.044 | 0.042 | 0.031 | 0.042 | 0.048 | 0.046 | 0.038 | 0.051 | 0.053 | 0.051 | 0.051 | 0.078 | 0.037 | 0.079 | 0.039 | 0.056 | 0.062 | 0.063 | 0.052 | 0.044 | 0.028 | 0.042 | 0.060 | 0.069 | 0.031 | 0.044 | 0.057 | 0.056 | 0.046 | 0.048 | 0.058 | 0.061 | 0.071 | 0.040 | 0.006 | 0.015 | 0.044 |
| Q4_10 | 0.075 | 0.077 | 0.079 | 0.090 | 0.075 | 0.061 | 0.030 | 0.081 | 0.073 | 0.000 | 0.072 | 0.096 | 0.114 | 0.055 | 0.081 | 0.076 | 0.081 | 0.103 | 0.087 | 0.066 | 0.025 | 0.062 | 0.044 | 0.043 | 0.055 | 0.053 | 0.051 | 0.069 | 0.066 | 0.060 | 0.053 | 0.083 | 0.071 | 0.103 | 0.067 | 0.074 | 0.103 | 0.106 | 0.082 | 0.067 | 0.047 | 0.060 | 0.066 | 0.075 | 0.042 | 0.048 | 0.059 | 0.058 | 0.046 | 0.050 | 0.057 | 0.089 | 0.075 | 0.024 | 0.018 | 0.021 | 0.066 |
| Q4_11 | 0.062 | 0.069 | 0.060 | 0.068 | 0.062 | 0.047 | 0.045 | 0.062 | 0.053 | 0.072 | 0.000 | 0.077 | 0.114 | 0.055 | 0.061 | 0.060 | 0.065 | 0.074 | 0.060 | 0.052 | 0.041 | 0.048 | 0.037 | 0.036 | 0.045 | 0.051 | 0.043 | 0.057 | 0.064 | 0.054 | 0.041 | 0.077 | 0.053 | 0.072 | 0.047 | 0.058 | 0.072 | 0.071 | 0.068 | 0.054 | 0.039 | 0.054 | 0.063 | 0.064 | 0.033 | 0.043 | 0.057 | 0.059 | 0.051 | 0.045 | 0.049 | 0.058 | 0.074 | 0.046 | 0.022 | 0.020 | 0.058 |
| Q4_12 | 0.063 | 0.069 | 0.058 | 0.083 | 0.072 | 0.053 | 0.049 | 0.067 | 0.053 | 0.096 | 0.077 | 0.000 | 0.121 | 0.085 | 0.064 | 0.071 | 0.063 | 0.081 | 0.062 | 0.059 | 0.054 | 0.051 | 0.053 | 0.041 | 0.056 | 0.069 | 0.054 | 0.068 | 0.083 | 0.051 | 0.054 | 0.092 | 0.068 | 0.094 | 0.058 | 0.079 | 0.099 | 0.083 | 0.086 | 0.066 | 0.057 | 0.074 | 0.070 | 0.084 | 0.063 | 0.051 | 0.070 | 0.057 | 0.064 | 0.053 | 0.060 | 0.057 | 0.067 | 0.046 | 0.007 | 0.013 | 0.058 |
| Q4_13 | 0.092 | 0.097 | 0.082 | 0.085 | 0.109 | 0.079 | 0.026 | 0.097 | 0.092 | 0.114 | 0.114 | 0.121 | 0.000 | 0.133 | 0.086 | 0.092 | 0.067 | 0.118 | 0.118 | 0.115 | 0.109 | 0.099 | 0.042 | 0.032 | 0.082 | 0.092 | 0.083 | 0.099 | 0.110 | 0.085 | 0.093 | 0.121 | 0.098 | 0.102 | 0.092 | 0.116 | 0.099 | 0.130 | 0.096 | 0.116 | 0.059 | 0.095 | 0.116 | 0.114 | 0.056 | 0.064 | 0.104 | 0.114 | 0.056 | 0.076 | 0.100 | 0.103 | 0.094 | 0.079 | -0.012 | -0.013 | 0.083 |
| Q4_14 | 0.064 | 0.050 | 0.043 | 0.048 | 0.060 | 0.045 | 0.047 | 0.060 | 0.051 | 0.055 | 0.055 | 0.085 | 0.133 | 0.000 | 0.071 | 0.069 | 0.058 | 0.067 | 0.059 | 0.055 | 0.040 | 0.058 | 0.034 | 0.025 | 0.049 | 0.046 | 0.043 | 0.058 | 0.049 | 0.059 | 0.032 | 0.102 | 0.071 | 0.086 | 0.063 | 0.066 | 0.079 | 0.086 | 0.081 | 0.066 | 0.058 | 0.047 | 0.058 | 0.047 | 0.028 | 0.033 | 0.046 | 0.057 | 0.053 | 0.033 | 0.055 | 0.070 | 0.053 | 0.051 | 0.022 | 0.006 | 0.060 |
| Q4_15 | 0.069 | 0.071 | 0.063 | 0.064 | 0.066 | 0.050 | 0.033 | 0.064 | 0.064 | 0.081 | 0.061 | 0.064 | 0.086 | 0.071 | 0.000 | 0.073 | 0.064 | 0.079 | 0.066 | 0.053 | 0.046 | 0.059 | 0.053 | 0.037 | 0.065 | 0.056 | 0.052 | 0.063 | 0.065 | 0.051 | 0.050 | 0.066 | 0.060 | 0.079 | 0.051 | 0.054 | 0.078 | 0.077 | 0.063 | 0.051 | 0.045 | 0.053 | 0.063 | 0.066 | 0.029 | 0.041 | 0.059 | 0.068 | 0.052 | 0.055 | 0.056 | 0.061 | 0.071 | 0.044 | 0.008 | 0.007 | 0.054 |
| Q4_16 | 0.066 | 0.064 | 0.059 | 0.066 | 0.062 | 0.054 | 0.046 | 0.059 | 0.054 | 0.076 | 0.060 | 0.071 | 0.092 | 0.069 | 0.073 | 0.000 | 0.072 | 0.073 | 0.063 | 0.049 | 0.040 | 0.059 | 0.038 | 0.031 | 0.066 | 0.053 | 0.045 | 0.062 | 0.060 | 0.053 | 0.047 | 0.095 | 0.071 | 0.084 | 0.061 | 0.043 | 0.085 | 0.072 | 0.066 | 0.050 | 0.048 | 0.049 | 0.057 | 0.054 | 0.017 | 0.037 | 0.048 | 0.052 | 0.028 | 0.035 | 0.054 | 0.066 | 0.053 | 0.036 | 0.004 | -0.004 | 0.060 |
| Q4_17 | 0.065 | 0.067 | 0.066 | 0.084 | 0.065 | 0.052 | 0.041 | 0.068 | 0.055 | 0.081 | 0.065 | 0.063 | 0.067 | 0.058 | 0.064 | 0.072 | 0.000 | 0.076 | 0.066 | 0.057 | 0.053 | 0.048 | 0.034 | 0.026 | 0.054 | 0.054 | 0.045 | 0.055 | 0.070 | 0.051 | 0.046 | 0.073 | 0.041 | 0.075 | 0.050 | 0.039 | 0.077 | 0.070 | 0.056 | 0.048 | 0.057 | 0.048 | 0.055 | 0.056 | 0.037 | 0.047 | 0.058 | 0.054 | 0.049 | 0.055 | 0.060 | 0.060 | 0.067 | 0.029 | -0.007 | 0.011 | 0.056 |
| Q4_18 | 0.079 | 0.074 | 0.072 | 0.086 | 0.068 | 0.066 | 0.033 | 0.077 | 0.066 | 0.103 | 0.074 | 0.081 | 0.118 | 0.067 | 0.079 | 0.073 | 0.076 | 0.000 | 0.072 | 0.067 | 0.058 | 0.060 | 0.046 | 0.051 | 0.069 | 0.058 | 0.061 | 0.075 | 0.088 | 0.054 | 0.054 | 0.084 | 0.052 | 0.084 | 0.049 | 0.051 | 0.084 | 0.082 | 0.061 | 0.058 | 0.053 | 0.064 | 0.073 | 0.071 | 0.031 | 0.054 | 0.060 | 0.057 | 0.054 | 0.050 | 0.062 | 0.076 | 0.092 | 0.040 | 0.000 | 0.024 | 0.072 |
| Q4_19 | 0.063 | 0.063 | 0.063 | 0.064 | 0.065 | 0.054 | 0.037 | 0.073 | 0.060 | 0.087 | 0.060 | 0.062 | 0.118 | 0.059 | 0.066 | 0.063 | 0.066 | 0.072 | 0.000 | 0.058 | 0.049 | 0.051 | 0.032 | 0.031 | 0.053 | 0.057 | 0.044 | 0.065 | 0.061 | 0.051 | 0.049 | 0.095 | 0.055 | 0.091 | 0.060 | 0.060 | 0.083 | 0.079 | 0.064 | 0.062 | 0.049 | 0.053 | 0.066 | 0.054 | 0.046 | 0.044 | 0.050 | 0.054 | 0.061 | 0.063 | 0.061 | 0.061 | 0.074 | 0.050 | 0.004 | 0.018 | 0.057 |
| Q5_1 | 0.052 | 0.066 | 0.054 | 0.056 | 0.057 | 0.040 | 0.036 | 0.050 | 0.051 | 0.066 | 0.052 | 0.059 | 0.115 | 0.055 | 0.053 | 0.049 | 0.057 | 0.067 | 0.058 | 0.000 | 0.044 | 0.057 | 0.047 | 0.036 | 0.056 | 0.044 | 0.051 | 0.057 | 0.066 | 0.054 | 0.052 | 0.078 | 0.038 | 0.049 | 0.027 | 0.062 | 0.075 | 0.052 | 0.048 | 0.056 | 0.028 | 0.048 | 0.057 | 0.049 | 0.024 | 0.033 | 0.047 | 0.042 | 0.043 | 0.034 | 0.051 | 0.051 | 0.069 | 0.041 | 0.011 | 0.014 | 0.052 |
| Q5_2 | 0.040 | 0.053 | 0.040 | 0.033 | 0.052 | 0.044 | 0.024 | 0.047 | 0.044 | 0.025 | 0.041 | 0.054 | 0.109 | 0.040 | 0.046 | 0.040 | 0.053 | 0.058 | 0.049 | 0.044 | 0.000 | 0.057 | 0.021 | 0.016 | 0.049 | 0.034 | 0.030 | 0.046 | 0.036 | 0.037 | 0.031 | 0.073 | 0.044 | 0.067 | 0.046 | 0.053 | 0.070 | 0.054 | 0.054 | 0.042 | 0.026 | 0.039 | 0.039 | 0.027 | 0.001 | 0.022 | 0.029 | 0.029 | 0.042 | 0.014 | 0.039 | 0.057 | 0.024 | 0.004 | 0.010 | 0.000 | 0.041 |
| Q5_3 | 0.045 | 0.059 | 0.044 | 0.051 | 0.052 | 0.046 | 0.030 | 0.052 | 0.042 | 0.062 | 0.048 | 0.051 | 0.099 | 0.058 | 0.059 | 0.059 | 0.048 | 0.060 | 0.051 | 0.057 | 0.057 | 0.000 | 0.052 | 0.043 | 0.061 | 0.044 | 0.053 | 0.056 | 0.057 | 0.051 | 0.047 | 0.086 | 0.049 | 0.058 | 0.041 | 0.043 | 0.065 | 0.053 | 0.055 | 0.051 | 0.038 | 0.046 | 0.056 | 0.055 | 0.020 | 0.033 | 0.042 | 0.046 | 0.044 | 0.040 | 0.050 | 0.054 | 0.048 | 0.023 | 0.007 | 0.004 | 0.051 |
| Q5_4 | 0.031 | 0.049 | 0.026 | 0.031 | 0.059 | 0.037 | 0.020 | 0.030 | 0.031 | 0.044 | 0.037 | 0.053 | 0.042 | 0.034 | 0.053 | 0.038 | 0.034 | 0.046 | 0.032 | 0.047 | 0.021 | 0.052 | 0.000 | 0.056 | 0.046 | 0.019 | 0.047 | 0.055 | 0.037 | 0.062 | 0.046 | 0.021 | 0.021 | 0.037 | 0.007 | 0.032 | 0.034 | 0.017 | 0.019 | 0.049 | 0.022 | 0.028 | 0.037 | 0.023 | 0.040 | 0.017 | 0.026 | 0.034 | 0.063 | 0.051 | 0.026 | 0.045 | 0.017 | 0.032 | 0.023 | 0.009 | 0.034 |
| Q5_5 | 0.036 | 0.046 | 0.035 | 0.034 | 0.048 | 0.033 | 0.009 | 0.030 | 0.042 | 0.043 | 0.036 | 0.041 | 0.032 | 0.025 | 0.037 | 0.031 | 0.026 | 0.051 | 0.031 | 0.036 | 0.016 | 0.043 | 0.056 | 0.000 | 0.041 | 0.014 | 0.043 | 0.051 | 0.037 | 0.060 | 0.045 | 0.025 | 0.032 | 0.036 | 0.015 | 0.040 | 0.020 | 0.021 | 0.021 | 0.043 | 0.012 | 0.017 | 0.039 | 0.022 | 0.030 | 0.011 | 0.015 | 0.031 | 0.039 | 0.034 | 0.029 | 0.050 | 0.008 | 0.022 | 0.024 | 0.015 | 0.034 |
| Q5_6 | 0.044 | 0.065 | 0.059 | 0.049 | 0.054 | 0.044 | 0.043 | 0.048 | 0.048 | 0.055 | 0.045 | 0.056 | 0.082 | 0.049 | 0.065 | 0.066 | 0.054 | 0.069 | 0.053 | 0.056 | 0.049 | 0.061 | 0.046 | 0.041 | 0.000 | 0.031 | 0.049 | 0.060 | 0.029 | 0.061 | 0.048 | 0.057 | 0.048 | 0.063 | 0.037 | 0.057 | 0.053 | 0.056 | 0.057 | 0.039 | 0.045 | 0.034 | 0.050 | 0.019 | 0.022 | 0.028 | 0.039 | 0.043 | 0.048 | 0.032 | 0.051 | 0.066 | 0.055 | 0.026 | 0.011 | 0.004 | 0.048 |
| Q5_7 | 0.042 | 0.043 | 0.040 | 0.040 | 0.048 | 0.040 | 0.065 | 0.043 | 0.046 | 0.053 | 0.051 | 0.069 | 0.092 | 0.046 | 0.056 | 0.053 | 0.054 | 0.058 | 0.057 | 0.044 | 0.034 | 0.044 | 0.019 | 0.014 | 0.031 | 0.000 | 0.035 | 0.056 | 0.051 | 0.049 | 0.030 | 0.083 | 0.062 | 0.084 | 0.051 | 0.057 | 0.072 | 0.080 | 0.073 | 0.062 | 0.043 | 0.050 | 0.055 | 0.049 | 0.043 | 0.034 | 0.037 | 0.038 | 0.030 | 0.028 | 0.050 | 0.048 | 0.049 | 0.027 | 0.016 | 0.004 | 0.038 |
| Q5_8 | 0.028 | 0.054 | 0.038 | 0.035 | 0.054 | 0.036 | 0.034 | 0.048 | 0.038 | 0.051 | 0.043 | 0.054 | 0.083 | 0.043 | 0.052 | 0.045 | 0.045 | 0.061 | 0.044 | 0.051 | 0.030 | 0.053 | 0.047 | 0.043 | 0.049 | 0.035 | 0.000 | 0.056 | 0.050 | 0.051 | 0.036 | 0.058 | 0.043 | 0.062 | 0.032 | 0.063 | 0.057 | 0.041 | 0.048 | 0.040 | 0.039 | 0.038 | 0.047 | 0.037 | 0.024 | 0.034 | 0.028 | 0.044 | 0.039 | 0.041 | 0.040 | 0.050 | 0.039 | 0.025 | 0.017 | 0.021 | 0.042 |
| Q5_9 | 0.055 | 0.066 | 0.056 | 0.056 | 0.066 | 0.052 | 0.049 | 0.053 | 0.051 | 0.069 | 0.057 | 0.068 | 0.099 | 0.058 | 0.063 | 0.062 | 0.055 | 0.075 | 0.065 | 0.057 | 0.046 | 0.056 | 0.055 | 0.051 | 0.060 | 0.056 | 0.056 | 0.000 | 0.074 | 0.054 | 0.049 | 0.088 | 0.062 | 0.069 | 0.050 | 0.059 | 0.071 | 0.067 | 0.063 | 0.058 | 0.053 | 0.056 | 0.058 | 0.051 | 0.032 | 0.043 | 0.047 | 0.045 | 0.036 | 0.042 | 0.056 | 0.056 | 0.064 | 0.034 | 0.000 | 0.014 | 0.059 |
| Q5_10 | 0.054 | 0.076 | 0.050 | 0.052 | 0.072 | 0.036 | 0.042 | 0.057 | 0.053 | 0.066 | 0.064 | 0.083 | 0.110 | 0.049 | 0.065 | 0.060 | 0.070 | 0.088 | 0.061 | 0.066 | 0.036 | 0.057 | 0.037 | 0.037 | 0.029 | 0.051 | 0.050 | 0.074 | 0.000 | 0.072 | 0.035 | 0.081 | 0.057 | 0.090 | 0.050 | 0.061 | 0.105 | 0.078 | 0.070 | 0.067 | 0.034 | 0.034 | 0.064 | 0.050 | 0.005 | 0.042 | 0.051 | 0.050 | 0.051 | 0.042 | 0.055 | 0.081 | 0.063 | 0.024 | -0.001 | 0.004 | 0.054 |
| Q5_11 | 0.052 | 0.060 | 0.058 | 0.049 | 0.058 | 0.042 | 0.041 | 0.046 | 0.051 | 0.060 | 0.054 | 0.051 | 0.085 | 0.059 | 0.051 | 0.053 | 0.051 | 0.054 | 0.051 | 0.054 | 0.037 | 0.051 | 0.062 | 0.060 | 0.061 | 0.049 | 0.051 | 0.054 | 0.072 | 0.000 | 0.054 | 0.083 | 0.052 | 0.049 | 0.029 | 0.055 | 0.064 | 0.055 | 0.052 | 0.042 | 0.056 | 0.047 | 0.061 | 0.046 | 0.049 | 0.046 | 0.046 | 0.049 | 0.069 | 0.048 | 0.055 | 0.056 | 0.057 | 0.056 | 0.036 | 0.030 | 0.050 |
| Q5_12 | 0.034 | 0.053 | 0.030 | 0.044 | 0.058 | 0.040 | 0.035 | 0.045 | 0.051 | 0.053 | 0.041 | 0.054 | 0.093 | 0.032 | 0.050 | 0.047 | 0.046 | 0.054 | 0.049 | 0.052 | 0.031 | 0.047 | 0.046 | 0.045 | 0.048 | 0.030 | 0.036 | 0.049 | 0.035 | 0.054 | 0.000 | 0.067 | 0.060 | 0.063 | 0.040 | 0.046 | 0.075 | 0.055 | 0.056 | 0.049 | 0.028 | 0.039 | 0.055 | 0.031 | 0.018 | 0.036 | 0.033 | 0.027 | 0.031 | 0.032 | 0.056 | 0.065 | 0.043 | 0.017 | 0.017 | -0.005 | 0.048 |
| Q6_1 | 0.077 | 0.086 | 0.090 | 0.071 | 0.073 | 0.060 | 0.048 | 0.069 | 0.078 | 0.083 | 0.077 | 0.092 | 0.121 | 0.102 | 0.066 | 0.095 | 0.073 | 0.084 | 0.095 | 0.078 | 0.073 | 0.086 | 0.021 | 0.025 | 0.057 | 0.083 | 0.058 | 0.088 | 0.081 | 0.083 | 0.067 | 0.000 | 0.093 | 0.102 | 0.090 | 0.095 | 0.098 | 0.113 | 0.100 | 0.072 | 0.066 | 0.075 | 0.100 | 0.084 | 0.025 | 0.057 | 0.066 | 0.077 | 0.024 | 0.046 | 0.100 | 0.081 | 0.071 | 0.063 | 0.010 | -0.009 | 0.100 |
| Q6_2 | 0.050 | 0.057 | 0.043 | 0.059 | 0.057 | 0.055 | 0.050 | 0.046 | 0.037 | 0.071 | 0.053 | 0.068 | 0.098 | 0.071 | 0.060 | 0.071 | 0.041 | 0.052 | 0.055 | 0.038 | 0.044 | 0.049 | 0.021 | 0.032 | 0.048 | 0.062 | 0.043 | 0.062 | 0.057 | 0.052 | 0.060 | 0.093 | 0.000 | 0.078 | 0.064 | 0.082 | 0.077 | 0.076 | 0.076 | 0.046 | 0.042 | 0.058 | 0.062 | 0.061 | 0.024 | 0.038 | 0.048 | 0.046 | 0.007 | 0.034 | 0.054 | 0.051 | 0.046 | 0.013 | -0.013 | -0.005 | 0.044 |
| Q6_3 | 0.069 | 0.066 | 0.069 | 0.085 | 0.074 | 0.060 | 0.053 | 0.073 | 0.079 | 0.103 | 0.072 | 0.094 | 0.102 | 0.086 | 0.079 | 0.084 | 0.075 | 0.084 | 0.091 | 0.049 | 0.067 | 0.058 | 0.037 | 0.036 | 0.063 | 0.084 | 0.062 | 0.069 | 0.090 | 0.049 | 0.063 | 0.102 | 0.078 | 0.000 | 0.091 | 0.087 | 0.098 | 0.099 | 0.090 | 0.069 | 0.047 | 0.077 | 0.088 | 0.079 | 0.048 | 0.054 | 0.067 | 0.075 | 0.033 | 0.041 | 0.076 | 0.077 | 0.070 | 0.013 | -0.022 | -0.001 | 0.069 |
| Q6_4 | 0.050 | 0.042 | 0.041 | 0.047 | 0.052 | 0.038 | 0.045 | 0.045 | 0.039 | 0.067 | 0.047 | 0.058 | 0.092 | 0.063 | 0.051 | 0.061 | 0.050 | 0.049 | 0.060 | 0.027 | 0.046 | 0.041 | 0.007 | 0.015 | 0.037 | 0.051 | 0.032 | 0.050 | 0.050 | 0.029 | 0.040 | 0.090 | 0.064 | 0.091 | 0.000 | 0.060 | 0.078 | 0.073 | 0.079 | 0.035 | 0.039 | 0.055 | 0.047 | 0.046 | 0.019 | 0.040 | 0.036 | 0.039 | -0.002 | 0.022 | 0.045 | 0.054 | 0.045 | 0.003 | -0.019 | -0.021 | 0.033 |
| Q6_5 | 0.050 | 0.060 | 0.065 | 0.065 | 0.052 | 0.034 | 0.024 | 0.052 | 0.056 | 0.074 | 0.058 | 0.079 | 0.116 | 0.066 | 0.054 | 0.043 | 0.039 | 0.051 | 0.060 | 0.062 | 0.053 | 0.043 | 0.032 | 0.040 | 0.057 | 0.057 | 0.063 | 0.059 | 0.061 | 0.055 | 0.046 | 0.095 | 0.082 | 0.087 | 0.060 | 0.000 | 0.081 | 0.063 | 0.070 | 0.057 | 0.055 | 0.055 | 0.078 | 0.074 | 0.027 | 0.049 | 0.046 | 0.055 | 0.028 | 0.038 | 0.064 | 0.063 | 0.059 | 0.035 | 0.002 | 0.016 | 0.063 |
| Q6_6 | 0.066 | 0.084 | 0.061 | 0.106 | 0.080 | 0.063 | 0.065 | 0.071 | 0.062 | 0.103 | 0.072 | 0.099 | 0.099 | 0.079 | 0.078 | 0.085 | 0.077 | 0.084 | 0.083 | 0.075 | 0.070 | 0.065 | 0.034 | 0.020 | 0.053 | 0.072 | 0.057 | 0.071 | 0.105 | 0.064 | 0.075 | 0.098 | 0.077 | 0.098 | 0.078 | 0.081 | 0.000 | 0.105 | 0.088 | 0.083 | 0.056 | 0.108 | 0.090 | 0.086 | 0.030 | 0.075 | 0.080 | 0.077 | 0.017 | 0.065 | 0.077 | 0.079 | 0.089 | 0.025 | -0.019 | -0.027 | 0.091 |
| Q6_7 | 0.065 | 0.076 | 0.065 | 0.071 | 0.076 | 0.054 | 0.043 | 0.067 | 0.063 | 0.106 | 0.071 | 0.083 | 0.130 | 0.086 | 0.077 | 0.072 | 0.070 | 0.082 | 0.079 | 0.052 | 0.054 | 0.053 | 0.017 | 0.021 | 0.056 | 0.080 | 0.041 | 0.067 | 0.078 | 0.055 | 0.055 | 0.113 | 0.076 | 0.099 | 0.073 | 0.063 | 0.105 | 0.000 | 0.086 | 0.067 | 0.057 | 0.082 | 0.075 | 0.070 | 0.046 | 0.055 | 0.065 | 0.052 | 0.037 | 0.047 | 0.059 | 0.069 | 0.070 | 0.027 | -0.013 | -0.011 | 0.071 |
| Q6_8 | 0.057 | 0.058 | 0.047 | 0.062 | 0.062 | 0.045 | 0.057 | 0.054 | 0.052 | 0.082 | 0.068 | 0.086 | 0.096 | 0.081 | 0.063 | 0.066 | 0.056 | 0.061 | 0.064 | 0.048 | 0.054 | 0.055 | 0.019 | 0.021 | 0.057 | 0.073 | 0.048 | 0.063 | 0.070 | 0.052 | 0.056 | 0.100 | 0.076 | 0.090 | 0.079 | 0.070 | 0.088 | 0.086 | 0.000 | 0.066 | 0.049 | 0.069 | 0.067 | 0.068 | 0.043 | 0.064 | 0.063 | 0.060 | 0.018 | 0.040 | 0.062 | 0.052 | 0.072 | 0.024 | -0.008 | 0.002 | 0.059 |
| Q6_9 | 0.042 | 0.052 | 0.045 | 0.055 | 0.050 | 0.037 | 0.040 | 0.051 | 0.044 | 0.067 | 0.054 | 0.066 | 0.116 | 0.066 | 0.051 | 0.050 | 0.048 | 0.058 | 0.062 | 0.056 | 0.042 | 0.051 | 0.049 | 0.043 | 0.039 | 0.062 | 0.040 | 0.058 | 0.067 | 0.042 | 0.049 | 0.072 | 0.046 | 0.069 | 0.035 | 0.057 | 0.083 | 0.067 | 0.066 | 0.000 | 0.067 | 0.065 | 0.067 | 0.066 | 0.055 | 0.050 | 0.064 | 0.056 | 0.068 | 0.052 | 0.059 | 0.059 | 0.066 | 0.071 | 0.038 | 0.036 | 0.063 |
| Q6_10 | 0.036 | 0.046 | 0.035 | 0.035 | 0.051 | 0.045 | 0.039 | 0.046 | 0.028 | 0.047 | 0.039 | 0.057 | 0.059 | 0.058 | 0.045 | 0.048 | 0.057 | 0.053 | 0.049 | 0.028 | 0.026 | 0.038 | 0.022 | 0.012 | 0.045 | 0.043 | 0.039 | 0.053 | 0.034 | 0.056 | 0.028 | 0.066 | 0.042 | 0.047 | 0.039 | 0.055 | 0.056 | 0.057 | 0.049 | 0.067 | 0.000 | 0.053 | 0.051 | 0.015 | 0.023 | 0.025 | 0.034 | 0.030 | 0.069 | 0.037 | 0.049 | 0.062 | 0.045 | 0.049 | 0.033 | 0.021 | 0.057 |
| Q6_11 | 0.047 | 0.061 | 0.046 | 0.045 | 0.058 | 0.043 | 0.045 | 0.057 | 0.042 | 0.060 | 0.054 | 0.074 | 0.095 | 0.047 | 0.053 | 0.049 | 0.048 | 0.064 | 0.053 | 0.048 | 0.039 | 0.046 | 0.028 | 0.017 | 0.034 | 0.050 | 0.038 | 0.056 | 0.034 | 0.047 | 0.039 | 0.075 | 0.058 | 0.077 | 0.055 | 0.055 | 0.108 | 0.082 | 0.069 | 0.065 | 0.053 | 0.000 | 0.059 | 0.038 | 0.024 | 0.030 | 0.047 | 0.039 | 0.039 | 0.028 | 0.048 | 0.062 | 0.048 | 0.026 | 0.023 | 0.021 | 0.058 |
| Q7_1 | 0.058 | 0.074 | 0.055 | 0.067 | 0.068 | 0.056 | 0.047 | 0.061 | 0.060 | 0.066 | 0.063 | 0.070 | 0.116 | 0.058 | 0.063 | 0.057 | 0.055 | 0.073 | 0.066 | 0.057 | 0.039 | 0.056 | 0.037 | 0.039 | 0.050 | 0.055 | 0.047 | 0.058 | 0.064 | 0.061 | 0.055 | 0.100 | 0.062 | 0.088 | 0.047 | 0.078 | 0.090 | 0.075 | 0.067 | 0.067 | 0.051 | 0.059 | 0.000 | 0.069 | 0.042 | 0.052 | 0.054 | 0.054 | 0.038 | 0.048 | 0.057 | 0.081 | 0.069 | 0.041 | 0.019 | 0.003 | 0.070 |
| Q7_2 | 0.050 | 0.066 | 0.042 | 0.057 | 0.060 | 0.051 | 0.050 | 0.059 | 0.069 | 0.075 | 0.064 | 0.084 | 0.114 | 0.047 | 0.066 | 0.054 | 0.056 | 0.071 | 0.054 | 0.049 | 0.027 | 0.055 | 0.023 | 0.022 | 0.019 | 0.049 | 0.037 | 0.051 | 0.050 | 0.046 | 0.031 | 0.084 | 0.061 | 0.079 | 0.046 | 0.074 | 0.086 | 0.070 | 0.068 | 0.066 | 0.015 | 0.038 | 0.069 | 0.000 | 0.058 | 0.045 | 0.033 | 0.029 | 0.044 | 0.030 | 0.047 | 0.075 | 0.045 | 0.030 | 0.029 | 0.010 | 0.068 |
| Q7_3 | 0.019 | 0.030 | 0.018 | 0.016 | 0.038 | 0.032 | 0.030 | 0.028 | 0.031 | 0.042 | 0.033 | 0.063 | 0.056 | 0.028 | 0.029 | 0.017 | 0.037 | 0.031 | 0.046 | 0.024 | 0.001 | 0.020 | 0.040 | 0.030 | 0.022 | 0.043 | 0.024 | 0.032 | 0.005 | 0.049 | 0.018 | 0.025 | 0.024 | 0.048 | 0.019 | 0.027 | 0.030 | 0.046 | 0.043 | 0.055 | 0.023 | 0.024 | 0.042 | 0.058 | 0.000 | 0.044 | 0.025 | 0.049 | 0.071 | 0.033 | 0.017 | 0.039 | 0.020 | 0.041 | 0.027 | 0.012 | 0.036 |
| Q7_4 | 0.038 | 0.046 | 0.036 | 0.056 | 0.053 | 0.038 | 0.039 | 0.037 | 0.044 | 0.048 | 0.043 | 0.051 | 0.064 | 0.033 | 0.041 | 0.037 | 0.047 | 0.054 | 0.044 | 0.033 | 0.022 | 0.033 | 0.017 | 0.011 | 0.028 | 0.034 | 0.034 | 0.043 | 0.042 | 0.046 | 0.036 | 0.057 | 0.038 | 0.054 | 0.040 | 0.049 | 0.075 | 0.055 | 0.064 | 0.050 | 0.025 | 0.030 | 0.052 | 0.045 | 0.044 | 0.000 | 0.037 | 0.036 | 0.037 | 0.044 | 0.046 | 0.063 | 0.052 | 0.029 | 0.016 | 0.022 | 0.053 |
| Q7_5 | 0.046 | 0.072 | 0.048 | 0.053 | 0.055 | 0.034 | 0.035 | 0.049 | 0.057 | 0.059 | 0.057 | 0.070 | 0.104 | 0.046 | 0.059 | 0.048 | 0.058 | 0.060 | 0.050 | 0.047 | 0.029 | 0.042 | 0.026 | 0.015 | 0.039 | 0.037 | 0.028 | 0.047 | 0.051 | 0.046 | 0.033 | 0.066 | 0.048 | 0.067 | 0.036 | 0.046 | 0.080 | 0.065 | 0.063 | 0.064 | 0.034 | 0.047 | 0.054 | 0.033 | 0.025 | 0.037 | 0.000 | 0.044 | 0.067 | 0.053 | 0.048 | 0.065 | 0.050 | 0.037 | 0.043 | 0.025 | 0.056 |
| Q7_6 | 0.049 | 0.058 | 0.053 | 0.054 | 0.057 | 0.038 | 0.025 | 0.055 | 0.056 | 0.058 | 0.059 | 0.057 | 0.114 | 0.057 | 0.068 | 0.052 | 0.054 | 0.057 | 0.054 | 0.042 | 0.029 | 0.046 | 0.034 | 0.031 | 0.043 | 0.038 | 0.044 | 0.045 | 0.050 | 0.049 | 0.027 | 0.077 | 0.046 | 0.075 | 0.039 | 0.055 | 0.077 | 0.052 | 0.060 | 0.056 | 0.030 | 0.039 | 0.054 | 0.029 | 0.049 | 0.036 | 0.044 | 0.000 | 0.058 | 0.042 | 0.043 | 0.071 | 0.050 | 0.045 | 0.033 | 0.040 | 0.045 |
| Q7_7 | 0.047 | 0.059 | 0.034 | 0.028 | 0.051 | 0.065 | 0.035 | 0.047 | 0.046 | 0.046 | 0.051 | 0.064 | 0.056 | 0.053 | 0.052 | 0.028 | 0.049 | 0.054 | 0.061 | 0.043 | 0.042 | 0.044 | 0.063 | 0.039 | 0.048 | 0.030 | 0.039 | 0.036 | 0.051 | 0.069 | 0.031 | 0.024 | 0.007 | 0.033 | -0.002 | 0.028 | 0.017 | 0.037 | 0.018 | 0.068 | 0.069 | 0.039 | 0.038 | 0.044 | 0.071 | 0.037 | 0.067 | 0.058 | 0.000 | 0.042 | 0.043 | 0.049 | 0.034 | 0.072 | 0.068 | 0.057 | 0.046 |
| Q7_8 | 0.034 | 0.055 | 0.042 | 0.038 | 0.053 | 0.033 | 0.018 | 0.052 | 0.048 | 0.050 | 0.045 | 0.053 | 0.076 | 0.033 | 0.055 | 0.035 | 0.055 | 0.050 | 0.063 | 0.034 | 0.014 | 0.040 | 0.051 | 0.034 | 0.032 | 0.028 | 0.041 | 0.042 | 0.042 | 0.048 | 0.032 | 0.046 | 0.034 | 0.041 | 0.022 | 0.038 | 0.065 | 0.047 | 0.040 | 0.052 | 0.037 | 0.028 | 0.048 | 0.030 | 0.033 | 0.044 | 0.053 | 0.042 | 0.042 | 0.000 | 0.057 | 0.067 | 0.059 | 0.048 | 0.038 | 0.026 | 0.046 |
| Q7_9 | 0.058 | 0.066 | 0.058 | 0.058 | 0.068 | 0.049 | 0.035 | 0.056 | 0.058 | 0.057 | 0.049 | 0.060 | 0.100 | 0.055 | 0.056 | 0.054 | 0.060 | 0.062 | 0.061 | 0.051 | 0.039 | 0.050 | 0.026 | 0.029 | 0.051 | 0.050 | 0.040 | 0.056 | 0.055 | 0.055 | 0.056 | 0.100 | 0.054 | 0.076 | 0.045 | 0.064 | 0.077 | 0.059 | 0.062 | 0.059 | 0.049 | 0.048 | 0.057 | 0.047 | 0.017 | 0.046 | 0.048 | 0.043 | 0.043 | 0.057 | 0.000 | 0.069 | 0.065 | 0.031 | 0.017 | 0.019 | 0.061 |
| Q7_10 | 0.075 | 0.086 | 0.069 | 0.081 | 0.071 | 0.065 | 0.051 | 0.064 | 0.061 | 0.089 | 0.058 | 0.057 | 0.103 | 0.070 | 0.061 | 0.066 | 0.060 | 0.076 | 0.061 | 0.051 | 0.057 | 0.054 | 0.045 | 0.050 | 0.066 | 0.048 | 0.050 | 0.056 | 0.081 | 0.056 | 0.065 | 0.081 | 0.051 | 0.077 | 0.054 | 0.063 | 0.079 | 0.069 | 0.052 | 0.059 | 0.062 | 0.062 | 0.081 | 0.075 | 0.039 | 0.063 | 0.065 | 0.071 | 0.049 | 0.067 | 0.069 | 0.000 | 0.086 | 0.035 | 0.006 | 0.023 | 0.081 |
| Q7_11 | 0.061 | 0.075 | 0.064 | 0.075 | 0.061 | 0.050 | 0.041 | 0.064 | 0.071 | 0.075 | 0.074 | 0.067 | 0.094 | 0.053 | 0.071 | 0.053 | 0.067 | 0.092 | 0.074 | 0.069 | 0.024 | 0.048 | 0.017 | 0.008 | 0.055 | 0.049 | 0.039 | 0.064 | 0.063 | 0.057 | 0.043 | 0.071 | 0.046 | 0.070 | 0.045 | 0.059 | 0.089 | 0.070 | 0.072 | 0.066 | 0.045 | 0.048 | 0.069 | 0.045 | 0.020 | 0.052 | 0.050 | 0.050 | 0.034 | 0.059 | 0.065 | 0.086 | 0.000 | 0.051 | 0.034 | 0.042 | 0.079 |
| Q7_12 | 0.044 | 0.034 | 0.029 | 0.017 | 0.033 | 0.029 | 0.012 | 0.025 | 0.040 | 0.024 | 0.046 | 0.046 | 0.079 | 0.051 | 0.044 | 0.036 | 0.029 | 0.040 | 0.050 | 0.041 | 0.004 | 0.023 | 0.032 | 0.022 | 0.026 | 0.027 | 0.025 | 0.034 | 0.024 | 0.056 | 0.017 | 0.063 | 0.013 | 0.013 | 0.003 | 0.035 | 0.025 | 0.027 | 0.024 | 0.071 | 0.049 | 0.026 | 0.041 | 0.030 | 0.041 | 0.029 | 0.037 | 0.045 | 0.072 | 0.048 | 0.031 | 0.035 | 0.051 | 0.000 | 0.076 | 0.044 | 0.031 |
| Q7_13 | 0.007 | 0.008 | 0.006 | 0.008 | -0.007 | 0.026 | 0.018 | 0.003 | 0.006 | 0.018 | 0.022 | 0.007 | -0.012 | 0.022 | 0.008 | 0.004 | -0.007 | 0.000 | 0.004 | 0.011 | 0.010 | 0.007 | 0.023 | 0.024 | 0.011 | 0.016 | 0.017 | 0.000 | -0.001 | 0.036 | 0.017 | 0.010 | -0.013 | -0.022 | -0.019 | 0.002 | -0.019 | -0.013 | -0.008 | 0.038 | 0.033 | 0.023 | 0.019 | 0.029 | 0.027 | 0.016 | 0.043 | 0.033 | 0.068 | 0.038 | 0.017 | 0.006 | 0.034 | 0.076 | 0.000 | 0.055 | 0.014 |
| Q7_14 | 0.011 | 0.020 | 0.019 | 0.006 | -0.001 | 0.031 | 0.002 | 0.010 | 0.015 | 0.021 | 0.020 | 0.013 | -0.013 | 0.006 | 0.007 | -0.004 | 0.011 | 0.024 | 0.018 | 0.014 | 0.000 | 0.004 | 0.009 | 0.015 | 0.004 | 0.004 | 0.021 | 0.014 | 0.004 | 0.030 | -0.005 | -0.009 | -0.005 | -0.001 | -0.021 | 0.016 | -0.027 | -0.011 | 0.002 | 0.036 | 0.021 | 0.021 | 0.003 | 0.010 | 0.012 | 0.022 | 0.025 | 0.040 | 0.057 | 0.026 | 0.019 | 0.023 | 0.042 | 0.044 | 0.055 | 0.000 | 0.012 |
| Q7_15 | 0.057 | 0.069 | 0.052 | 0.062 | 0.069 | 0.057 | 0.048 | 0.049 | 0.044 | 0.066 | 0.058 | 0.058 | 0.083 | 0.060 | 0.054 | 0.060 | 0.056 | 0.072 | 0.057 | 0.052 | 0.041 | 0.051 | 0.034 | 0.034 | 0.048 | 0.038 | 0.042 | 0.059 | 0.054 | 0.050 | 0.048 | 0.100 | 0.044 | 0.069 | 0.033 | 0.063 | 0.091 | 0.071 | 0.059 | 0.063 | 0.057 | 0.058 | 0.070 | 0.068 | 0.036 | 0.053 | 0.056 | 0.045 | 0.046 | 0.046 | 0.061 | 0.081 | 0.079 | 0.031 | 0.014 | 0.012 | 0.000 |
# What % of PC correlations are higher than PPM?
a <- sum(round(corr.pc$rho - corr.ppm, 3) > 0)/2
b <- sum(round(corr.pc$rho - corr.ppm, 3) < 0)/2
prop.diff <- a/(a+b)
prop.diff[1] 0.9868173# Is this difference significantly different than .5?
X <- table(c(rep(0,a),rep(1,b)))
prop.test(x=X, alternative = 'greater')
1-sample proportions test with continuity correction
data: X, null probability 0.5
X-squared = 1508.2, df = 1, p-value < 2.2e-16
alternative hypothesis: true p is greater than 0.5
95 percent confidence interval:
0.9808562 1.0000000
sample estimates:
p
0.9868173 Eigenvalues and Scree Plot
evalues<-eigen(corr.pc$rho)$values
evalues [1] 2.627313e+01 3.761010e+00 2.952063e+00 2.210781e+00 1.460550e+00
[6] 1.397493e+00 1.274058e+00 1.101728e+00 1.011097e+00 9.481728e-01
[11] 8.631385e-01 7.519192e-01 7.257157e-01 7.148329e-01 6.489492e-01
[16] 6.317037e-01 6.070564e-01 5.773193e-01 5.627900e-01 5.426212e-01
[21] 5.001011e-01 4.759799e-01 4.542633e-01 4.423228e-01 4.201005e-01
[26] 4.067098e-01 3.823340e-01 3.695824e-01 3.251477e-01 3.064071e-01
[31] 2.924240e-01 2.856197e-01 2.675262e-01 2.658989e-01 2.568959e-01
[36] 2.417409e-01 2.170894e-01 2.100147e-01 1.954760e-01 1.827397e-01
[41] 1.693641e-01 1.613067e-01 1.502327e-01 1.461206e-01 1.426707e-01
[46] 1.223941e-01 1.080088e-01 9.494902e-02 8.224543e-02 7.330500e-02
[51] 6.150090e-02 5.520100e-02 4.169916e-02 3.892286e-02 2.619869e-02
[56] 1.138168e-02 9.990371e-11plotnScree(nScree(evalues, model="factors"), main="Scree Plot & Parallel Analysis")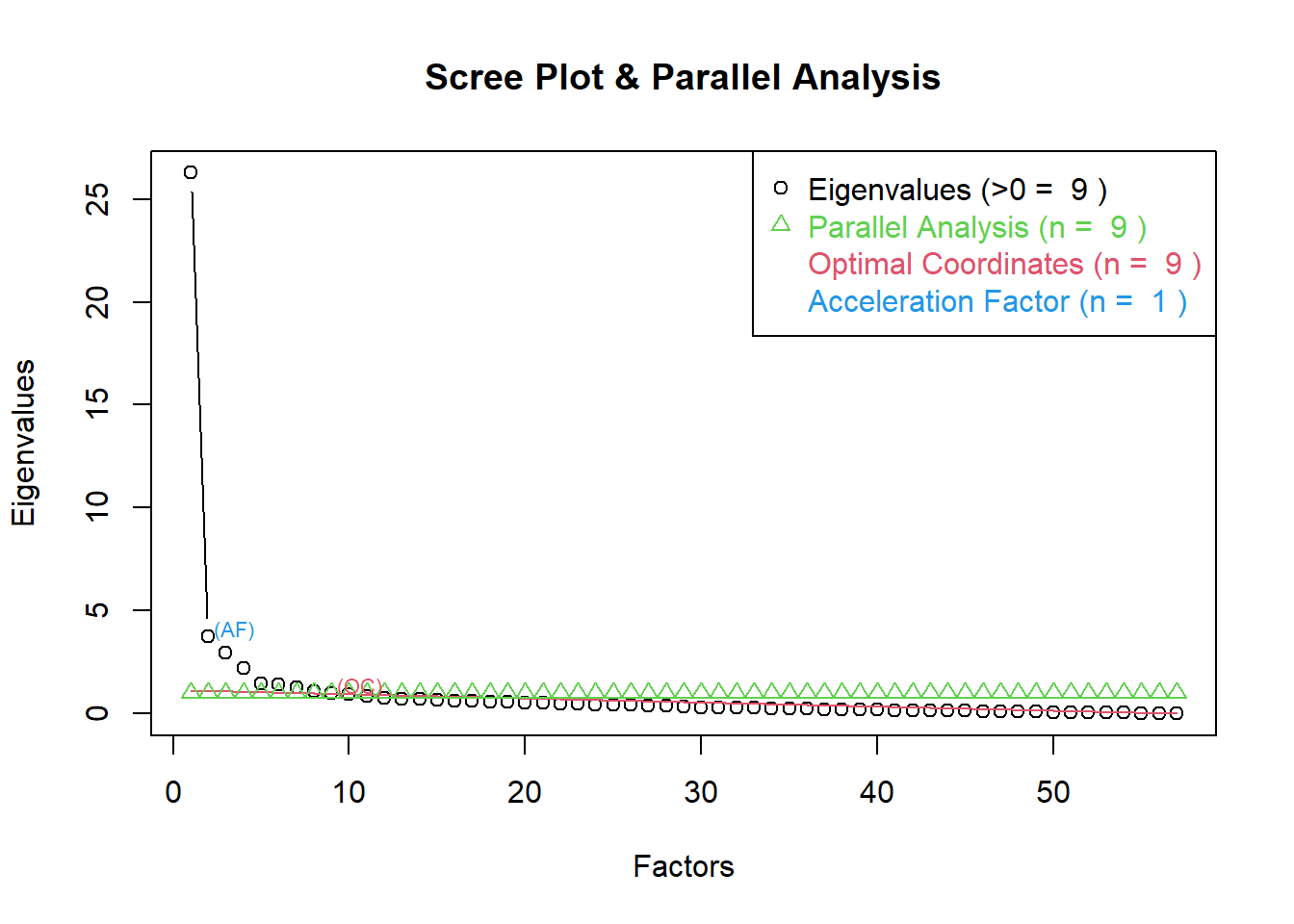
psych::fa.parallel(dat,main="Parallel Analysis",
fm = "pa",cor = "cov")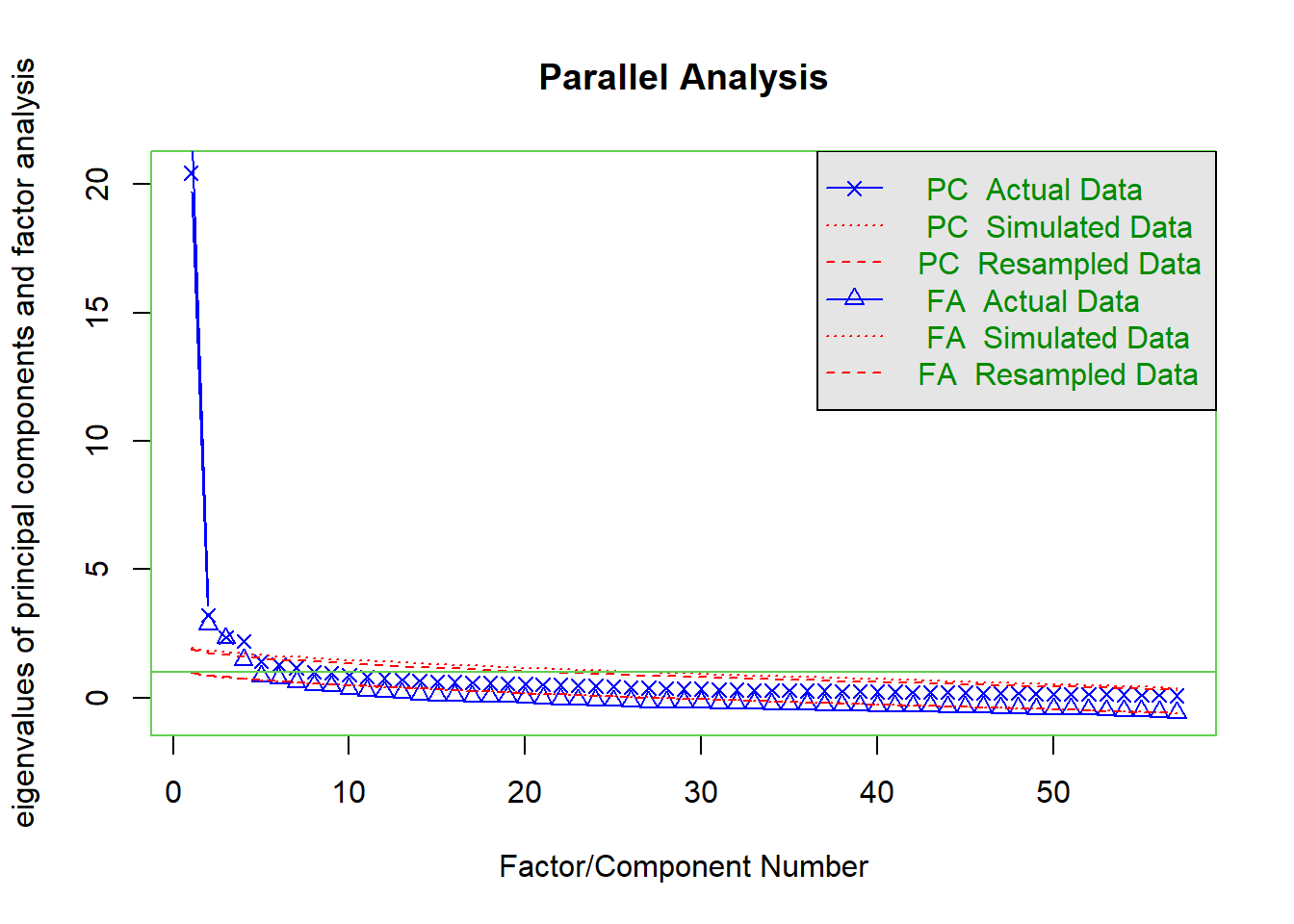
Parallel analysis suggests that the number of factors = 6 and the number of components = 4 psych::fa.parallel(corr.pc$rho,main="Parallel Analysis",
n.obs = nrow(mydata), fm = "pa", cor = "cov")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.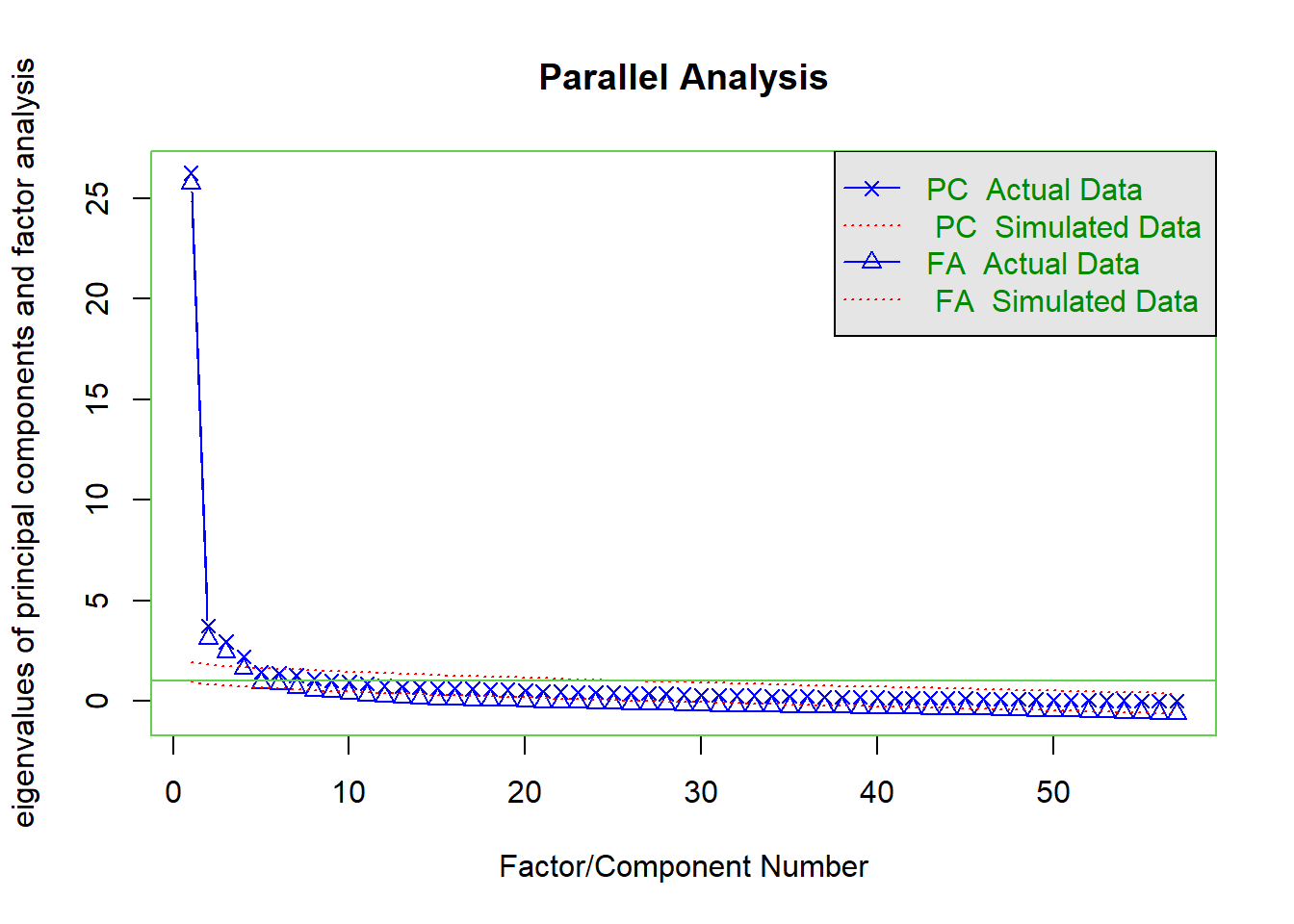
Parallel analysis suggests that the number of factors = 7 and the number of components = 4 Note that the warning for the “estimated weights for the factor scores…” is not particularly impacting because we do not intend to obtain factor scores.
Factor Extraction
Two factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 2, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 2, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA1 PA2 h2 u2 com
Q4_1 0.41 0.33 0.47 0.53 1.9
Q4_2 0.43 0.36 0.55 0.45 1.9
Q4_3 0.36 0.39 0.48 0.52 2.0
Q4_4 0.33 0.48 0.57 0.43 1.8
Q4_5 0.52 0.27 0.54 0.46 1.5
Q4_6 0.25 0.48 0.47 0.53 1.5
Q4_7 0.05 0.55 0.34 0.66 1.0
Q4_8 0.58 0.18 0.52 0.48 1.2
Q4_9 0.38 0.32 0.43 0.57 1.9
Q4_10 0.36 0.42 0.53 0.47 2.0
Q4_11 0.46 0.37 0.60 0.40 1.9
Q4_12 0.70 0.07 0.56 0.44 1.0
Q4_13 0.80 -0.06 0.58 0.42 1.0
Q4_14 0.51 0.28 0.55 0.45 1.6
Q4_15 0.54 0.29 0.60 0.40 1.5
Q4_16 0.57 0.19 0.52 0.48 1.2
Q4_17 0.54 0.19 0.48 0.52 1.2
Q4_18 0.51 0.35 0.65 0.35 1.8
Q4_19 0.45 0.39 0.61 0.39 2.0
Q5_1 0.23 0.50 0.47 0.53 1.4
Q5_2 0.21 0.48 0.43 0.57 1.4
Q5_3 0.30 0.42 0.45 0.55 1.8
Q5_4 -0.16 0.77 0.43 0.57 1.1
Q5_5 -0.22 0.75 0.37 0.63 1.2
Q5_6 0.12 0.64 0.53 0.47 1.1
Q5_7 0.43 0.28 0.43 0.57 1.7
Q5_8 0.12 0.61 0.49 0.51 1.1
Q5_9 0.44 0.38 0.58 0.42 2.0
Q5_10 0.34 0.36 0.42 0.58 2.0
Q5_11 0.25 0.51 0.50 0.50 1.4
Q5_12 0.20 0.53 0.47 0.53 1.3
Q6_1 0.97 -0.32 0.59 0.41 1.2
Q6_2 0.93 -0.27 0.58 0.42 1.2
Q6_3 0.93 -0.26 0.59 0.41 1.2
Q6_4 0.91 -0.32 0.50 0.50 1.3
Q6_5 0.45 0.13 0.30 0.70 1.2
Q6_6 0.94 -0.17 0.68 0.32 1.1
Q6_7 0.95 -0.16 0.72 0.28 1.1
Q6_8 0.95 -0.21 0.65 0.35 1.1
Q6_9 0.26 0.35 0.31 0.69 1.9
Q6_10 0.28 0.37 0.37 0.63 1.9
Q6_11 0.38 0.41 0.54 0.46 2.0
Q7_1 0.43 0.43 0.64 0.36 2.0
Q7_2 0.21 0.57 0.54 0.46 1.3
Q7_3 -0.27 0.88 0.50 0.50 1.2
Q7_4 0.10 0.60 0.45 0.55 1.1
Q7_5 0.10 0.70 0.60 0.40 1.0
Q7_6 0.13 0.63 0.52 0.48 1.1
Q7_7 -0.40 1.06 0.67 0.33 1.3
Q7_8 -0.01 0.71 0.49 0.51 1.0
Q7_9 0.38 0.42 0.55 0.45 2.0
Q7_10 0.52 0.30 0.60 0.40 1.6
Q7_11 0.13 0.63 0.54 0.46 1.1
Q7_12 -0.15 0.74 0.41 0.59 1.1
Q7_13 -0.36 0.65 0.21 0.79 1.6
Q7_14 -0.46 0.91 0.43 0.57 1.5
Q7_15 0.40 0.36 0.50 0.50 2.0
PA1 PA2
SS loadings 14.67 14.42
Proportion Var 0.26 0.25
Cumulative Var 0.26 0.51
Proportion Explained 0.50 0.50
Cumulative Proportion 0.50 1.00
With factor correlations of
PA1 PA2
PA1 1.00 0.72
PA2 0.72 1.00
Mean item complexity = 1.5
Test of the hypothesis that 2 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1483 and the objective function was 48.88
The root mean square of the residuals (RMSR) is 0.07
The df corrected root mean square of the residuals is 0.07
Fit based upon off diagonal values = 0.98print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA2
Q4_1 0.409 0.329
Q4_2 0.434 0.363
Q4_3 0.356 0.387
Q4_4 0.332 0.480
Q4_5 0.518
Q4_6 0.480
Q4_7 0.549
Q4_8 0.577
Q4_9 0.383 0.319
Q4_10 0.361 0.420
Q4_11 0.459 0.372
Q4_12 0.696
Q4_13 0.802
Q4_14 0.512
Q4_15 0.541
Q4_16 0.567
Q4_17 0.543
Q4_18 0.514 0.350
Q4_19 0.452 0.388
Q5_1 0.501
Q5_2 0.484
Q5_3 0.302 0.421
Q5_4 0.766
Q5_5 0.751
Q5_6 0.638
Q5_7 0.428
Q5_8 0.609
Q5_9 0.438 0.385
Q5_10 0.343 0.359
Q5_11 0.508
Q5_12 0.528
Q6_1 0.969 -0.320
Q6_2 0.930
Q6_3 0.930
Q6_4 0.906 -0.323
Q6_5 0.447
Q6_6 0.944
Q6_7 0.955
Q6_8 0.948
Q6_9 0.345
Q6_10 0.370
Q6_11 0.382 0.410
Q7_1 0.430 0.431
Q7_2 0.570
Q7_3 0.879
Q7_4 0.596
Q7_5 0.701
Q7_6 0.627
Q7_7 -0.402 1.058
Q7_8 0.707
Q7_9 0.381 0.421
Q7_10 0.524 0.305
Q7_11 0.633
Q7_12 0.738
Q7_13 -0.358 0.646
Q7_14 -0.460 0.909
Q7_15 0.398 0.361
PA1 PA2
SS loadings 13.541 13.290
Proportion Var 0.238 0.233
Cumulative Var 0.238 0.471fit <- psych::fa(r = cov.dat, nfactors = 2, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 2, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA2 h2 u2 H2 U2
Q4_1 0.48 0.11 0.31 0.44 0.41 0.59
Q4_2 0.49 0.10 0.32 0.35 0.48 0.52
Q4_3 0.42 0.15 0.29 0.41 0.42 0.58
Q4_4 0.37 0.25 0.33 0.34 0.49 0.51
Q4_5 0.55 0.08 0.37 0.40 0.48 0.52
Q4_6 0.33 0.36 0.41 0.55 0.42 0.58
Q4_7 0.08 0.48 0.29 0.59 0.33 0.67
Q4_8 0.67 -0.04 0.42 0.44 0.48 0.52
Q4_9 0.53 0.12 0.39 0.63 0.38 0.62
Q4_10 0.35 0.20 0.27 0.35 0.43 0.57
Q4_11 0.58 0.16 0.49 0.42 0.54 0.46
Q4_12 0.79 -0.08 0.54 0.57 0.49 0.51
Q4_13 0.72 -0.20 0.36 0.41 0.46 0.54
Q4_14 0.53 0.13 0.39 0.41 0.49 0.51
Q4_15 0.63 0.07 0.47 0.38 0.55 0.45
Q4_16 0.60 0.06 0.41 0.48 0.46 0.54
Q4_17 0.65 -0.01 0.41 0.52 0.44 0.56
Q4_18 0.56 0.08 0.39 0.29 0.58 0.42
Q4_19 0.58 0.18 0.51 0.43 0.55 0.45
Q5_1 0.32 0.35 0.38 0.52 0.42 0.58
Q5_2 0.22 0.50 0.46 0.62 0.43 0.57
Q5_3 0.36 0.37 0.46 0.64 0.42 0.58
Q5_4 -0.11 0.81 0.53 0.69 0.44 0.56
Q5_5 -0.19 0.77 0.41 0.66 0.39 0.61
Q5_6 0.15 0.54 0.42 0.40 0.52 0.48
Q5_7 0.39 0.25 0.35 0.54 0.39 0.61
Q5_8 0.17 0.60 0.53 0.58 0.48 0.52
Q5_9 0.48 0.30 0.52 0.47 0.53 0.47
Q5_10 0.29 0.26 0.26 0.44 0.37 0.63
Q5_11 0.35 0.34 0.40 0.50 0.44 0.56
Q5_12 0.22 0.46 0.41 0.50 0.45 0.55
Q6_1 0.70 -0.26 0.30 0.41 0.42 0.58
Q6_2 0.73 -0.20 0.37 0.46 0.44 0.56
Q6_3 0.71 -0.19 0.35 0.50 0.41 0.59
Q6_4 0.70 -0.23 0.32 0.54 0.37 0.63
Q6_5 0.44 0.08 0.25 0.81 0.24 0.76
Q6_6 0.71 -0.17 0.36 0.31 0.53 0.47
Q6_7 0.78 -0.17 0.45 0.31 0.59 0.41
Q6_8 0.79 -0.22 0.44 0.38 0.53 0.47
Q6_9 0.41 0.26 0.39 1.11 0.26 0.74
Q6_10 0.32 0.26 0.28 0.55 0.34 0.66
Q6_11 0.38 0.36 0.47 0.47 0.50 0.50
Q7_1 0.43 0.31 0.47 0.35 0.57 0.43
Q7_2 0.20 0.44 0.37 0.37 0.50 0.50
Q7_3 -0.20 0.85 0.52 0.50 0.51 0.49
Q7_4 0.15 0.52 0.40 0.56 0.42 0.58
Q7_5 0.19 0.55 0.49 0.40 0.55 0.45
Q7_6 0.19 0.49 0.41 0.44 0.48 0.52
Q7_7 -0.23 0.92 0.59 0.44 0.58 0.42
Q7_8 0.10 0.51 0.34 0.43 0.44 0.56
Q7_9 0.46 0.29 0.49 0.50 0.50 0.50
Q7_10 0.59 0.13 0.47 0.42 0.52 0.48
Q7_11 0.18 0.43 0.33 0.38 0.47 0.53
Q7_12 -0.02 0.63 0.38 0.66 0.37 0.63
Q7_13 -0.20 0.52 0.16 0.82 0.17 0.83
Q7_14 -0.32 0.82 0.40 0.57 0.41 0.59
Q7_15 0.43 0.21 0.36 0.46 0.44 0.56
PA1 PA2
SS loadings 13.13 9.53
Proportion Var 0.26 0.19
Cumulative Var 0.26 0.45
Proportion Explained 0.58 0.42
Cumulative Proportion 0.58 1.00
Standardized loadings (pattern matrix)
item PA1 PA2 h2 u2
Q4_1 1 0.55 0.13 0.41 0.59
Q4_2 2 0.60 0.12 0.48 0.52
Q4_3 3 0.50 0.18 0.42 0.58
Q4_4 4 0.45 0.31 0.49 0.51
Q4_5 5 0.63 0.09 0.48 0.52
Q4_6 6 0.34 0.37 0.42 0.58
Q4_7 7 0.08 0.51 0.33 0.67
Q4_8 8 0.72 -0.04 0.48 0.52
Q4_9 9 0.53 0.11 0.38 0.62
Q4_10 10 0.45 0.26 0.43 0.57
Q4_11 11 0.61 0.16 0.54 0.46
Q4_12 12 0.75 -0.08 0.49 0.51
Q4_13 13 0.82 -0.23 0.46 0.54
Q4_14 14 0.59 0.15 0.49 0.51
Q4_15 15 0.69 0.08 0.55 0.45
Q4_16 16 0.64 0.06 0.46 0.54
Q4_17 17 0.67 -0.01 0.44 0.56
Q4_18 18 0.69 0.10 0.58 0.42
Q4_19 19 0.60 0.19 0.55 0.45
Q5_1 20 0.33 0.37 0.42 0.58
Q5_2 21 0.22 0.48 0.43 0.57
Q5_3 22 0.35 0.35 0.42 0.58
Q5_4 23 -0.10 0.73 0.44 0.56
Q5_5 24 -0.19 0.74 0.39 0.61
Q5_6 25 0.16 0.59 0.52 0.48
Q5_7 26 0.41 0.26 0.39 0.61
Q5_8 27 0.16 0.57 0.48 0.52
Q5_9 28 0.48 0.30 0.53 0.47
Q5_10 29 0.35 0.31 0.37 0.63
Q5_11 30 0.36 0.36 0.44 0.56
Q5_12 31 0.23 0.49 0.45 0.55
Q6_1 32 0.83 -0.31 0.42 0.58
Q6_2 33 0.80 -0.22 0.44 0.56
Q6_3 34 0.77 -0.21 0.41 0.59
Q6_4 35 0.76 -0.25 0.37 0.63
Q6_5 36 0.43 0.08 0.24 0.76
Q6_6 37 0.86 -0.20 0.53 0.47
Q6_7 38 0.89 -0.20 0.59 0.41
Q6_8 39 0.88 -0.24 0.53 0.47
Q6_9 40 0.33 0.22 0.26 0.74
Q6_10 41 0.35 0.28 0.34 0.66
Q6_11 42 0.39 0.37 0.50 0.50
Q7_1 43 0.47 0.34 0.57 0.43
Q7_2 44 0.24 0.52 0.50 0.50
Q7_3 45 -0.19 0.84 0.51 0.49
Q7_4 46 0.15 0.53 0.42 0.58
Q7_5 47 0.20 0.58 0.55 0.45
Q7_6 48 0.20 0.54 0.48 0.52
Q7_7 49 -0.23 0.90 0.58 0.42
Q7_8 50 0.12 0.58 0.44 0.56
Q7_9 51 0.47 0.29 0.50 0.50
Q7_10 52 0.62 0.13 0.52 0.48
Q7_11 53 0.22 0.51 0.47 0.53
Q7_12 54 -0.02 0.62 0.37 0.63
Q7_13 55 -0.21 0.53 0.17 0.83
Q7_14 56 -0.32 0.83 0.41 0.59
Q7_15 57 0.48 0.23 0.44 0.56
PA1 PA2
SS loadings 14.74 10.70
Proportion Var 0.26 0.19
Cumulative Var 0.26 0.45
Cum. factor Var 0.58 1.00
With factor correlations of
PA1 PA2
PA1 1.00 0.71
PA2 0.71 1.00
Mean item complexity = 1.3
Test of the hypothesis that 2 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1483 and the objective function was 15.97
The root mean square of the residuals (RMSR) is 0.06
The df corrected root mean square of the residuals is 0.06
Fit based upon off diagonal values = 0.97
Measures of factor score adequacy
PA1 PA2
Correlation of (regression) scores with factors 0.98 0.97
Multiple R square of scores with factors 0.97 0.95
Minimum correlation of possible factor scores 0.94 0.90print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA2
Q4_1 0.476
Q4_2 0.492
Q4_3 0.420
Q4_4 0.368
Q4_5 0.554
Q4_6 0.332 0.358
Q4_7 0.478
Q4_8 0.671
Q4_9 0.534
Q4_10 0.355
Q4_11 0.583
Q4_12 0.789
Q4_13 0.720
Q4_14 0.526
Q4_15 0.629
Q4_16 0.597
Q4_17 0.647
Q4_18 0.564
Q4_19 0.578
Q5_1 0.316 0.354
Q5_2 0.498
Q5_3 0.362 0.370
Q5_4 0.806
Q5_5 0.766
Q5_6 0.539
Q5_7 0.390
Q5_8 0.602
Q5_9 0.482
Q5_10
Q5_11 0.346 0.341
Q5_12 0.463
Q6_1 0.698
Q6_2 0.734
Q6_3 0.712
Q6_4 0.701
Q6_5 0.441
Q6_6 0.705
Q6_7 0.782
Q6_8 0.795
Q6_9 0.405
Q6_10 0.319
Q6_11 0.383 0.361
Q7_1 0.427 0.310
Q7_2 0.444
Q7_3 0.849
Q7_4 0.521
Q7_5 0.554
Q7_6 0.493
Q7_7 0.918
Q7_8 0.507
Q7_9 0.463
Q7_10 0.589
Q7_11 0.430
Q7_12 0.633
Q7_13 0.522
Q7_14 -0.315 0.817
Q7_15 0.431
PA1 PA2
SS loadings 12.383 8.785
Proportion Var 0.217 0.154
Cumulative Var 0.217 0.371Three factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 3, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 3, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA1 PA2 PA3 h2 u2 com
Q4_1 1.00 -0.07 -0.20 0.66 0.34 1.1
Q4_2 0.99 -0.03 -0.17 0.72 0.28 1.1
Q4_3 0.98 -0.01 -0.24 0.66 0.34 1.1
Q4_4 0.85 0.14 -0.19 0.69 0.31 1.2
Q4_5 0.68 0.02 0.09 0.58 0.42 1.0
Q4_6 0.29 0.38 0.08 0.47 0.53 2.0
Q4_7 0.04 0.54 0.03 0.35 0.65 1.0
Q4_8 0.97 -0.20 -0.02 0.68 0.32 1.1
Q4_9 0.74 0.03 -0.07 0.50 0.50 1.0
Q4_10 0.74 0.13 -0.09 0.60 0.40 1.1
Q4_11 0.72 0.10 0.01 0.65 0.35 1.0
Q4_12 0.43 -0.05 0.41 0.56 0.44 2.0
Q4_13 0.60 -0.25 0.41 0.59 0.41 2.2
Q4_14 0.48 0.12 0.21 0.55 0.45 1.5
Q4_15 0.77 0.00 0.06 0.67 0.33 1.0
Q4_16 0.66 -0.05 0.15 0.56 0.44 1.1
Q4_17 0.75 -0.09 0.08 0.55 0.45 1.1
Q4_18 1.04 -0.06 -0.12 0.83 0.17 1.0
Q4_19 0.73 0.11 0.00 0.67 0.33 1.0
Q5_1 0.38 0.36 0.00 0.48 0.52 2.0
Q5_2 0.07 0.48 0.17 0.44 0.56 1.3
Q5_3 0.21 0.36 0.17 0.45 0.55 2.1
Q5_4 0.00 0.75 -0.15 0.44 0.56 1.1
Q5_5 0.02 0.72 -0.22 0.37 0.63 1.2
Q5_6 0.16 0.58 0.03 0.54 0.46 1.2
Q5_7 -0.05 0.35 0.45 0.48 0.52 1.9
Q5_8 -0.01 0.63 0.13 0.52 0.48 1.1
Q5_9 0.21 0.34 0.30 0.59 0.41 2.6
Q5_10 0.21 0.30 0.21 0.43 0.57 2.6
Q5_11 0.22 0.44 0.11 0.50 0.50 1.6
Q5_12 0.04 0.54 0.17 0.49 0.51 1.2
Q6_1 -0.06 -0.21 0.98 0.67 0.33 1.1
Q6_2 -0.21 -0.10 1.05 0.72 0.28 1.1
Q6_3 -0.19 -0.10 1.03 0.72 0.28 1.1
Q6_4 -0.18 -0.17 1.00 0.61 0.39 1.1
Q6_5 0.03 0.16 0.42 0.32 0.68 1.3
Q6_6 0.01 -0.09 0.91 0.74 0.26 1.0
Q6_7 0.08 -0.10 0.88 0.76 0.24 1.0
Q6_8 -0.08 -0.09 0.98 0.74 0.26 1.0
Q6_9 0.02 0.37 0.24 0.33 0.67 1.7
Q6_10 0.02 0.39 0.26 0.39 0.61 1.7
Q6_11 -0.04 0.47 0.40 0.59 0.41 2.0
Q7_1 0.11 0.43 0.35 0.66 0.34 2.1
Q7_2 0.04 0.58 0.18 0.56 0.44 1.2
Q7_3 -0.24 0.98 -0.11 0.57 0.43 1.2
Q7_4 -0.07 0.65 0.14 0.48 0.52 1.1
Q7_5 0.11 0.67 0.04 0.61 0.39 1.1
Q7_6 0.04 0.63 0.11 0.54 0.46 1.1
Q7_7 -0.05 1.05 -0.35 0.68 0.32 1.2
Q7_8 0.10 0.67 -0.06 0.50 0.50 1.1
Q7_9 0.29 0.33 0.20 0.55 0.45 2.6
Q7_10 0.38 0.19 0.28 0.60 0.40 2.4
Q7_11 0.25 0.54 -0.02 0.54 0.46 1.4
Q7_12 -0.06 0.76 -0.10 0.43 0.57 1.0
Q7_13 -0.23 0.72 -0.20 0.24 0.76 1.4
Q7_14 -0.21 0.97 -0.31 0.46 0.54 1.3
Q7_15 0.28 0.28 0.22 0.50 0.50 2.9
PA1 PA2 PA3
SS loadings 11.98 11.37 8.44
Proportion Var 0.21 0.20 0.15
Cumulative Var 0.21 0.41 0.56
Proportion Explained 0.38 0.36 0.27
Cumulative Proportion 0.38 0.73 1.00
With factor correlations of
PA1 PA2 PA3
PA1 1.00 0.74 0.73
PA2 0.74 1.00 0.68
PA3 0.73 0.68 1.00
Mean item complexity = 1.4
Test of the hypothesis that 3 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1428 and the objective function was 44.36
The root mean square of the residuals (RMSR) is 0.05
The df corrected root mean square of the residuals is 0.06
Fit based upon off diagonal values = 0.99print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA2 PA3
Q4_1 0.997
Q4_2 0.990
Q4_3 0.982
Q4_4 0.854
Q4_5 0.680
Q4_6 0.380
Q4_7 0.543
Q4_8 0.970
Q4_9 0.737
Q4_10 0.739
Q4_11 0.721
Q4_12 0.431 0.411
Q4_13 0.595 0.415
Q4_14 0.476
Q4_15 0.770
Q4_16 0.664
Q4_17 0.747
Q4_18 1.039
Q4_19 0.727
Q5_1 0.379 0.361
Q5_2 0.479
Q5_3 0.365
Q5_4 0.754
Q5_5 0.724
Q5_6 0.583
Q5_7 0.349 0.446
Q5_8 0.635
Q5_9 0.342 0.302
Q5_10 0.304
Q5_11 0.440
Q5_12 0.539
Q6_1 0.985
Q6_2 1.050
Q6_3 1.033
Q6_4 1.000
Q6_5 0.415
Q6_6 0.914
Q6_7 0.880
Q6_8 0.979
Q6_9 0.369
Q6_10 0.395
Q6_11 0.473 0.395
Q7_1 0.431 0.350
Q7_2 0.581
Q7_3 0.981
Q7_4 0.648
Q7_5 0.673
Q7_6 0.632
Q7_7 1.051 -0.349
Q7_8 0.670
Q7_9 0.335
Q7_10 0.381
Q7_11 0.540
Q7_12 0.762
Q7_13 0.721
Q7_14 0.969 -0.312
Q7_15
PA1 PA2 PA3
SS loadings 11.797 11.310 8.941
Proportion Var 0.207 0.198 0.157
Cumulative Var 0.207 0.405 0.562fit <- psych::fa(r = cov.dat, nfactors = 3, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 3, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA2 PA1 PA3 h2 u2 H2 U2
Q4_1 -0.05 0.80 -0.17 0.42 0.33 0.57 0.43
Q4_2 -0.05 0.78 -0.15 0.43 0.25 0.63 0.37
Q4_3 0.00 0.76 -0.20 0.40 0.30 0.57 0.43
Q4_4 0.14 0.63 -0.16 0.39 0.27 0.59 0.41
Q4_5 -0.01 0.60 0.06 0.40 0.37 0.52 0.48
Q4_6 0.35 0.27 0.08 0.41 0.56 0.42 0.58
Q4_7 0.50 0.06 -0.01 0.29 0.59 0.33 0.67
Q4_8 -0.20 0.87 -0.03 0.52 0.34 0.61 0.39
Q4_9 -0.02 0.76 -0.09 0.47 0.54 0.46 0.54
Q4_10 0.11 0.55 -0.11 0.31 0.31 0.50 0.50
Q4_11 0.05 0.71 -0.01 0.55 0.37 0.60 0.40
Q4_12 -0.11 0.51 0.36 0.54 0.57 0.49 0.51
Q4_13 -0.25 0.54 0.29 0.36 0.41 0.47 0.53
Q4_14 0.09 0.47 0.13 0.40 0.40 0.50 0.50
Q4_15 -0.04 0.72 0.03 0.52 0.32 0.62 0.38
Q4_16 -0.03 0.62 0.09 0.44 0.45 0.49 0.51
Q4_17 -0.13 0.73 0.06 0.46 0.47 0.49 0.51
Q4_18 -0.07 0.83 -0.11 0.50 0.18 0.73 0.27
Q4_19 0.06 0.75 -0.04 0.58 0.36 0.62 0.38
Q5_1 0.32 0.35 -0.01 0.39 0.51 0.43 0.57
Q5_2 0.54 0.06 0.13 0.46 0.61 0.43 0.57
Q5_3 0.38 0.22 0.15 0.46 0.64 0.42 0.58
Q5_4 0.85 -0.06 -0.13 0.53 0.69 0.44 0.56
Q5_5 0.79 -0.06 -0.20 0.41 0.66 0.38 0.62
Q5_6 0.56 0.10 0.02 0.42 0.40 0.52 0.48
Q5_7 0.32 -0.01 0.37 0.38 0.50 0.43 0.57
Q5_8 0.67 -0.02 0.13 0.55 0.56 0.50 0.50
Q5_9 0.33 0.21 0.28 0.53 0.46 0.53 0.47
Q5_10 0.27 0.16 0.13 0.26 0.44 0.37 0.63
Q5_11 0.34 0.24 0.11 0.40 0.50 0.44 0.56
Q5_12 0.51 0.05 0.14 0.41 0.49 0.46 0.54
Q6_1 -0.17 -0.04 0.75 0.39 0.32 0.55 0.45
Q6_2 -0.08 -0.16 0.89 0.55 0.28 0.66 0.34
Q6_3 -0.07 -0.18 0.88 0.53 0.33 0.62 0.38
Q6_4 -0.12 -0.13 0.84 0.46 0.40 0.54 0.46
Q6_5 0.13 0.06 0.37 0.27 0.80 0.25 0.75
Q6_6 -0.09 0.03 0.69 0.43 0.24 0.64 0.36
Q6_7 -0.10 0.10 0.71 0.51 0.25 0.67 0.33
Q6_8 -0.12 0.00 0.82 0.54 0.27 0.67 0.33
Q6_9 0.30 0.14 0.25 0.39 1.10 0.26 0.74
Q6_10 0.30 0.07 0.23 0.29 0.54 0.35 0.65
Q6_11 0.44 0.00 0.34 0.51 0.43 0.54 0.46
Q7_1 0.35 0.13 0.29 0.48 0.33 0.59 0.41
Q7_2 0.49 0.05 0.12 0.37 0.36 0.51 0.49
Q7_3 0.92 -0.20 -0.10 0.54 0.49 0.53 0.47
Q7_4 0.58 -0.03 0.13 0.42 0.54 0.44 0.56
Q7_5 0.57 0.14 0.03 0.50 0.40 0.55 0.45
Q7_6 0.53 0.06 0.09 0.41 0.43 0.49 0.51
Q7_7 0.94 -0.03 -0.27 0.59 0.44 0.57 0.43
Q7_8 0.52 0.12 -0.04 0.34 0.43 0.44 0.56
Q7_9 0.29 0.31 0.18 0.49 0.50 0.50 0.50
Q7_10 0.12 0.37 0.26 0.47 0.43 0.52 0.48
Q7_11 0.42 0.20 -0.02 0.33 0.38 0.47 0.53
Q7_12 0.67 -0.02 -0.05 0.39 0.66 0.37 0.63
Q7_13 0.56 -0.16 -0.11 0.17 0.81 0.17 0.83
Q7_14 0.87 -0.19 -0.21 0.40 0.57 0.42 0.58
Q7_15 0.21 0.28 0.18 0.36 0.46 0.44 0.56
PA2 PA1 PA3
SS loadings 9.48 9.40 5.85
Proportion Var 0.19 0.19 0.12
Cumulative Var 0.19 0.37 0.49
Proportion Explained 0.38 0.38 0.24
Cumulative Proportion 0.38 0.76 1.00
Standardized loadings (pattern matrix)
item PA2 PA1 PA3 h2 u2
Q4_1 1 -0.05 0.92 -0.20 0.57 0.43
Q4_2 2 -0.06 0.96 -0.18 0.63 0.37
Q4_3 3 0.00 0.91 -0.24 0.57 0.43
Q4_4 4 0.17 0.78 -0.20 0.59 0.41
Q4_5 5 -0.01 0.68 0.07 0.52 0.48
Q4_6 6 0.36 0.27 0.08 0.42 0.58
Q4_7 7 0.53 0.06 -0.01 0.33 0.67
Q4_8 8 -0.21 0.94 -0.03 0.61 0.39
Q4_9 9 -0.02 0.76 -0.09 0.46 0.54
Q4_10 10 0.14 0.69 -0.13 0.50 0.50
Q4_11 11 0.05 0.75 -0.01 0.60 0.40
Q4_12 12 -0.11 0.49 0.35 0.49 0.51
Q4_13 13 -0.29 0.61 0.33 0.47 0.53
Q4_14 14 0.10 0.52 0.14 0.50 0.50
Q4_15 15 -0.04 0.79 0.04 0.62 0.38
Q4_16 16 -0.03 0.66 0.09 0.49 0.51
Q4_17 17 -0.13 0.75 0.06 0.49 0.51
Q4_18 18 -0.08 1.01 -0.13 0.73 0.27
Q4_19 19 0.06 0.77 -0.05 0.62 0.38
Q5_1 20 0.34 0.37 -0.01 0.43 0.57
Q5_2 21 0.52 0.06 0.12 0.43 0.57
Q5_3 22 0.37 0.21 0.14 0.42 0.58
Q5_4 23 0.76 -0.05 -0.11 0.44 0.56
Q5_5 24 0.77 -0.06 -0.19 0.38 0.62
Q5_6 25 0.62 0.11 0.02 0.52 0.48
Q5_7 26 0.34 -0.01 0.39 0.43 0.57
Q5_8 27 0.63 -0.02 0.12 0.50 0.50
Q5_9 28 0.33 0.21 0.28 0.53 0.47
Q5_10 29 0.32 0.19 0.16 0.37 0.63
Q5_11 30 0.36 0.26 0.12 0.44 0.56
Q5_12 31 0.53 0.05 0.14 0.46 0.54
Q6_1 32 -0.20 -0.05 0.89 0.55 0.45
Q6_2 33 -0.09 -0.18 0.98 0.66 0.34
Q6_3 34 -0.08 -0.19 0.95 0.62 0.38
Q6_4 35 -0.13 -0.14 0.90 0.54 0.46
Q6_5 36 0.13 0.06 0.36 0.25 0.75
Q6_6 37 -0.11 0.04 0.84 0.64 0.36
Q6_7 38 -0.11 0.11 0.81 0.67 0.33
Q6_8 39 -0.14 -0.01 0.90 0.67 0.33
Q6_9 40 0.25 0.12 0.21 0.26 0.74
Q6_10 41 0.33 0.07 0.26 0.35 0.65
Q6_11 42 0.45 0.00 0.35 0.54 0.46
Q7_1 43 0.39 0.14 0.32 0.59 0.41
Q7_2 44 0.57 0.05 0.14 0.51 0.49
Q7_3 45 0.91 -0.20 -0.10 0.53 0.47
Q7_4 46 0.59 -0.03 0.13 0.44 0.56
Q7_5 47 0.61 0.14 0.03 0.55 0.45
Q7_6 48 0.57 0.07 0.10 0.49 0.51
Q7_7 49 0.93 -0.03 -0.27 0.57 0.43
Q7_8 50 0.59 0.14 -0.04 0.44 0.56
Q7_9 51 0.29 0.32 0.18 0.50 0.50
Q7_10 52 0.12 0.40 0.28 0.52 0.48
Q7_11 53 0.50 0.24 -0.02 0.47 0.53
Q7_12 54 0.65 -0.02 -0.05 0.37 0.63
Q7_13 55 0.57 -0.16 -0.12 0.17 0.83
Q7_14 56 0.88 -0.20 -0.22 0.42 0.58
Q7_15 57 0.23 0.30 0.20 0.44 0.56
PA2 PA1 PA3
SS loadings 10.65 10.55 6.57
Proportion Var 0.19 0.19 0.12
Cumulative Var 0.19 0.37 0.49
Cum. factor Var 0.38 0.76 1.00
With factor correlations of
PA2 PA1 PA3
PA2 1.00 0.74 0.65
PA1 0.74 1.00 0.72
PA3 0.65 0.72 1.00
Mean item complexity = 1.4
Test of the hypothesis that 3 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1428 and the objective function was 12.51
The root mean square of the residuals (RMSR) is 0.05
The df corrected root mean square of the residuals is 0.05
Fit based upon off diagonal values = 0.98
Measures of factor score adequacy
PA2 PA1 PA3
Correlation of (regression) scores with factors 0.98 0.98 0.98
Multiple R square of scores with factors 0.95 0.97 0.95
Minimum correlation of possible factor scores 0.90 0.93 0.90print(fit$loadings, cutoff=0.3)
Loadings:
PA2 PA1 PA3
Q4_1 0.796
Q4_2 0.784
Q4_3 0.758
Q4_4 0.633
Q4_5 0.596
Q4_6 0.349
Q4_7 0.497
Q4_8 0.875
Q4_9 0.764
Q4_10 0.548
Q4_11 0.712
Q4_12 0.512 0.364
Q4_13 0.538
Q4_14 0.470
Q4_15 0.724
Q4_16 0.616
Q4_17 0.727
Q4_18 0.827
Q4_19 0.748
Q5_1 0.322 0.352
Q5_2 0.540
Q5_3 0.382
Q5_4 0.846
Q5_5 0.792
Q5_6 0.560
Q5_7 0.322 0.368
Q5_8 0.667
Q5_9 0.327
Q5_10
Q5_11 0.341
Q5_12 0.507
Q6_1 0.751
Q6_2 0.893
Q6_3 0.881
Q6_4 0.837
Q6_5 0.372
Q6_6 0.688
Q6_7 0.709
Q6_8 0.815
Q6_9 0.301
Q6_10 0.300
Q6_11 0.439 0.343
Q7_1 0.354
Q7_2 0.486
Q7_3 0.923
Q7_4 0.581
Q7_5 0.574
Q7_6 0.528
Q7_7 0.938
Q7_8 0.516
Q7_9 0.313
Q7_10 0.374
Q7_11 0.422
Q7_12 0.670
Q7_13 0.564
Q7_14 0.867
Q7_15
PA2 PA1 PA3
SS loadings 9.482 9.219 6.036
Proportion Var 0.166 0.162 0.106
Cumulative Var 0.166 0.328 0.434Four factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 4, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 4, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA1 PA3 PA2 PA4 h2 u2 com
Q4_1 0.99 -0.20 -0.09 0.02 0.66 0.34 1.1
Q4_2 0.99 -0.18 -0.06 0.04 0.72 0.28 1.1
Q4_3 0.98 -0.25 -0.02 0.01 0.66 0.34 1.1
Q4_4 0.86 -0.19 0.13 0.00 0.69 0.31 1.1
Q4_5 0.67 0.08 -0.09 0.14 0.59 0.41 1.2
Q4_6 0.27 0.06 0.15 0.30 0.48 0.52 2.6
Q4_7 0.03 0.02 0.33 0.28 0.35 0.65 2.0
Q4_8 0.98 -0.01 -0.13 -0.10 0.68 0.32 1.1
Q4_9 0.76 -0.06 0.13 -0.13 0.52 0.48 1.1
Q4_10 0.75 -0.09 0.15 -0.04 0.61 0.39 1.1
Q4_11 0.74 0.02 0.13 -0.05 0.66 0.34 1.1
Q4_12 0.44 0.42 0.03 -0.12 0.57 0.43 2.2
Q4_13 0.60 0.42 -0.17 -0.10 0.59 0.41 2.1
Q4_14 0.48 0.21 0.10 0.02 0.56 0.44 1.5
Q4_15 0.76 0.06 -0.06 0.08 0.67 0.33 1.0
Q4_16 0.64 0.13 -0.23 0.24 0.59 0.41 1.6
Q4_17 0.76 0.09 -0.01 -0.12 0.56 0.44 1.1
Q4_18 1.03 -0.13 -0.11 0.06 0.83 0.17 1.1
Q4_19 0.74 0.01 0.12 -0.02 0.67 0.33 1.1
Q5_1 0.35 -0.02 0.08 0.36 0.50 0.50 2.1
Q5_2 0.02 0.13 0.00 0.64 0.55 0.45 1.1
Q5_3 0.14 0.12 -0.15 0.69 0.61 0.39 1.2
Q5_4 -0.09 -0.23 0.10 0.93 0.67 0.33 1.2
Q5_5 -0.06 -0.30 0.09 0.90 0.59 0.41 1.3
Q5_6 0.09 -0.03 0.00 0.80 0.71 0.29 1.0
Q5_7 -0.07 0.43 0.11 0.30 0.49 0.51 2.0
Q5_8 -0.08 0.08 0.08 0.75 0.65 0.35 1.1
Q5_9 0.16 0.27 -0.05 0.52 0.66 0.34 1.8
Q5_10 0.19 0.19 0.03 0.35 0.45 0.55 2.1
Q5_11 0.21 0.10 0.25 0.24 0.50 0.50 3.3
Q5_12 0.01 0.15 0.21 0.43 0.51 0.49 1.8
Q6_1 -0.07 0.98 -0.19 -0.03 0.67 0.33 1.1
Q6_2 -0.22 1.05 -0.16 0.07 0.72 0.28 1.1
Q6_3 -0.20 1.03 -0.16 0.07 0.72 0.28 1.1
Q6_4 -0.18 1.00 -0.19 0.02 0.61 0.39 1.1
Q6_5 0.04 0.43 0.18 -0.04 0.32 0.68 1.4
Q6_6 0.02 0.93 -0.02 -0.11 0.75 0.25 1.0
Q6_7 0.09 0.90 0.02 -0.17 0.78 0.22 1.1
Q6_8 -0.07 1.00 0.03 -0.18 0.77 0.23 1.1
Q6_9 0.06 0.28 0.53 -0.23 0.41 0.59 1.9
Q6_10 0.04 0.28 0.37 0.02 0.40 0.60 1.9
Q6_11 -0.03 0.40 0.38 0.11 0.59 0.41 2.2
Q7_1 0.11 0.35 0.28 0.18 0.66 0.34 2.7
Q7_2 0.06 0.19 0.49 0.10 0.57 0.43 1.4
Q7_3 -0.22 -0.09 0.89 0.11 0.60 0.40 1.2
Q7_4 -0.04 0.16 0.63 0.01 0.51 0.49 1.1
Q7_5 0.14 0.07 0.74 -0.10 0.68 0.32 1.1
Q7_6 0.07 0.13 0.66 -0.05 0.59 0.41 1.1
Q7_7 -0.02 -0.33 0.96 0.12 0.72 0.28 1.3
Q7_8 0.13 -0.04 0.70 -0.05 0.55 0.45 1.1
Q7_9 0.30 0.20 0.27 0.07 0.56 0.44 2.9
Q7_10 0.39 0.28 0.17 0.02 0.60 0.40 2.3
Q7_11 0.28 0.00 0.57 -0.05 0.58 0.42 1.5
Q7_12 -0.03 -0.07 0.79 -0.04 0.49 0.51 1.0
Q7_13 -0.19 -0.17 0.82 -0.12 0.32 0.68 1.3
Q7_14 -0.19 -0.30 0.83 0.17 0.48 0.52 1.5
Q7_15 0.29 0.22 0.23 0.05 0.50 0.50 2.9
PA1 PA3 PA2 PA4
SS loadings 11.90 8.39 7.74 5.64
Proportion Var 0.21 0.15 0.14 0.10
Cumulative Var 0.21 0.36 0.49 0.59
Proportion Explained 0.35 0.25 0.23 0.17
Cumulative Proportion 0.35 0.60 0.83 1.00
With factor correlations of
PA1 PA3 PA2 PA4
PA1 1.00 0.74 0.70 0.67
PA3 0.74 1.00 0.64 0.63
PA2 0.70 0.64 1.00 0.68
PA4 0.67 0.63 0.68 1.00
Mean item complexity = 1.5
Test of the hypothesis that 4 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1374 and the objective function was 42.03
The root mean square of the residuals (RMSR) is 0.04
The df corrected root mean square of the residuals is 0.05
Fit based upon off diagonal values = 0.99print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA3 PA2 PA4
Q4_1 0.994
Q4_2 0.987
Q4_3 0.983
Q4_4 0.863
Q4_5 0.668
Q4_6 0.300
Q4_7 0.325
Q4_8 0.976
Q4_9 0.758
Q4_10 0.752
Q4_11 0.736
Q4_12 0.445 0.424
Q4_13 0.599 0.418
Q4_14 0.481
Q4_15 0.763
Q4_16 0.640
Q4_17 0.761
Q4_18 1.033
Q4_19 0.737
Q5_1 0.355 0.363
Q5_2 0.639
Q5_3 0.693
Q5_4 0.929
Q5_5 0.899
Q5_6 0.796
Q5_7 0.433 0.304
Q5_8 0.755
Q5_9 0.519
Q5_10 0.352
Q5_11
Q5_12 0.427
Q6_1 0.984
Q6_2 1.045
Q6_3 1.028
Q6_4 0.996
Q6_5 0.427
Q6_6 0.926
Q6_7 0.901
Q6_8 1.003
Q6_9 0.531
Q6_10 0.370
Q6_11 0.404 0.378
Q7_1 0.349
Q7_2 0.491
Q7_3 0.890
Q7_4 0.625
Q7_5 0.740
Q7_6 0.660
Q7_7 -0.333 0.957
Q7_8 0.703
Q7_9 0.301
Q7_10 0.390
Q7_11 0.571
Q7_12 0.791
Q7_13 0.817
Q7_14 0.832
Q7_15
PA1 PA3 PA2 PA4
SS loadings 11.819 9.051 7.625 5.243
Proportion Var 0.207 0.159 0.134 0.092
Cumulative Var 0.207 0.366 0.500 0.592fit <- psych::fa(r = cov.dat, nfactors = 4, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 4, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA3 PA2 PA4 h2 u2 H2 U2
Q4_1 0.81 -0.18 -0.09 0.03 0.43 0.32 0.57 0.43
Q4_2 0.79 -0.16 -0.07 0.01 0.43 0.25 0.63 0.37
Q4_3 0.77 -0.21 -0.02 0.02 0.40 0.30 0.57 0.43
Q4_4 0.64 -0.17 0.11 0.03 0.40 0.27 0.59 0.41
Q4_5 0.60 0.06 -0.09 0.08 0.41 0.37 0.52 0.48
Q4_6 0.27 0.08 0.13 0.26 0.41 0.55 0.43 0.57
Q4_7 0.06 -0.01 0.29 0.25 0.29 0.59 0.32 0.68
Q4_8 0.89 -0.03 -0.14 -0.09 0.52 0.34 0.61 0.39
Q4_9 0.78 -0.10 0.09 -0.14 0.48 0.53 0.48 0.52
Q4_10 0.56 -0.11 0.12 -0.01 0.32 0.31 0.51 0.49
Q4_11 0.73 -0.01 0.09 -0.07 0.55 0.36 0.60 0.40
Q4_12 0.52 0.37 0.01 -0.16 0.55 0.56 0.49 0.51
Q4_13 0.54 0.29 -0.17 -0.13 0.36 0.41 0.47 0.53
Q4_14 0.48 0.13 0.10 -0.02 0.40 0.40 0.50 0.50
Q4_15 0.73 0.03 -0.08 0.03 0.52 0.32 0.62 0.38
Q4_16 0.62 0.08 -0.21 0.19 0.46 0.42 0.52 0.48
Q4_17 0.74 0.05 -0.03 -0.14 0.46 0.47 0.50 0.50
Q4_18 0.84 -0.12 -0.10 0.02 0.50 0.18 0.73 0.27
Q4_19 0.76 -0.05 0.08 -0.04 0.58 0.36 0.62 0.38
Q5_1 0.35 -0.01 0.08 0.28 0.40 0.50 0.44 0.56
Q5_2 0.05 0.13 0.04 0.60 0.55 0.52 0.51 0.49
Q5_3 0.20 0.14 -0.14 0.61 0.58 0.51 0.53 0.47
Q5_4 -0.11 -0.16 0.09 0.97 0.78 0.45 0.64 0.36
Q5_5 -0.10 -0.23 0.08 0.91 0.63 0.44 0.59 0.41
Q5_6 0.08 0.01 0.01 0.66 0.54 0.28 0.65 0.35
Q5_7 -0.02 0.38 0.10 0.26 0.39 0.50 0.44 0.56
Q5_8 -0.05 0.13 0.08 0.71 0.67 0.45 0.60 0.40
Q5_9 0.20 0.29 -0.05 0.42 0.58 0.41 0.58 0.42
Q5_10 0.16 0.14 0.03 0.28 0.27 0.43 0.39 0.61
Q5_11 0.25 0.12 0.21 0.16 0.40 0.50 0.44 0.56
Q5_12 0.05 0.14 0.21 0.35 0.42 0.48 0.47 0.53
Q6_1 -0.05 0.76 -0.16 -0.04 0.39 0.32 0.55 0.45
Q6_2 -0.17 0.91 -0.13 0.03 0.55 0.28 0.66 0.34
Q6_3 -0.19 0.90 -0.14 0.05 0.53 0.33 0.62 0.38
Q6_4 -0.14 0.85 -0.17 0.02 0.46 0.40 0.54 0.46
Q6_5 0.07 0.38 0.19 -0.07 0.28 0.78 0.26 0.74
Q6_6 0.03 0.70 -0.03 -0.09 0.43 0.24 0.64 0.36
Q6_7 0.10 0.72 0.01 -0.15 0.52 0.24 0.68 0.32
Q6_8 0.00 0.84 0.01 -0.18 0.56 0.26 0.68 0.32
Q6_9 0.16 0.27 0.58 -0.31 0.52 0.97 0.35 0.65
Q6_10 0.07 0.24 0.32 -0.02 0.31 0.53 0.37 0.63
Q6_11 0.00 0.35 0.37 0.08 0.51 0.43 0.55 0.45
Q7_1 0.14 0.29 0.23 0.14 0.48 0.34 0.59 0.41
Q7_2 0.05 0.13 0.39 0.12 0.38 0.36 0.51 0.49
Q7_3 -0.20 -0.10 0.86 0.12 0.59 0.43 0.58 0.42
Q7_4 -0.02 0.13 0.56 0.04 0.44 0.52 0.46 0.54
Q7_5 0.15 0.04 0.66 -0.07 0.56 0.34 0.63 0.37
Q7_6 0.08 0.10 0.59 -0.05 0.46 0.38 0.55 0.45
Q7_7 -0.02 -0.28 0.90 0.11 0.65 0.38 0.63 0.37
Q7_8 0.13 -0.03 0.57 -0.04 0.39 0.38 0.50 0.50
Q7_9 0.32 0.18 0.23 0.06 0.49 0.50 0.50 0.50
Q7_10 0.38 0.27 0.11 0.00 0.47 0.42 0.52 0.48
Q7_11 0.22 -0.02 0.43 0.00 0.35 0.35 0.50 0.50
Q7_12 -0.01 -0.05 0.73 -0.03 0.46 0.59 0.44 0.56
Q7_13 -0.15 -0.12 0.73 -0.14 0.27 0.71 0.27 0.73
Q7_14 -0.19 -0.22 0.75 0.18 0.43 0.55 0.44 0.56
Q7_15 0.28 0.18 0.17 0.04 0.36 0.46 0.44 0.56
PA1 PA3 PA2 PA4
SS loadings 9.51 5.94 6.32 4.73
Proportion Var 0.19 0.12 0.12 0.09
Cumulative Var 0.19 0.30 0.43 0.52
Proportion Explained 0.36 0.22 0.24 0.18
Cumulative Proportion 0.36 0.58 0.82 1.00
Standardized loadings (pattern matrix)
item PA1 PA3 PA2 PA4 h2 u2
Q4_1 1 0.93 -0.21 -0.10 0.04 0.57 0.43
Q4_2 2 0.97 -0.19 -0.08 0.01 0.63 0.37
Q4_3 3 0.92 -0.25 -0.02 0.02 0.57 0.43
Q4_4 4 0.79 -0.21 0.13 0.04 0.59 0.41
Q4_5 5 0.68 0.06 -0.10 0.09 0.52 0.48
Q4_6 6 0.28 0.08 0.13 0.26 0.43 0.57
Q4_7 7 0.07 -0.01 0.31 0.26 0.32 0.68
Q4_8 8 0.95 -0.04 -0.15 -0.09 0.61 0.39
Q4_9 9 0.78 -0.10 0.09 -0.14 0.48 0.52
Q4_10 10 0.71 -0.14 0.15 -0.01 0.51 0.49
Q4_11 11 0.76 -0.01 0.10 -0.07 0.60 0.40
Q4_12 12 0.50 0.35 0.01 -0.15 0.49 0.51
Q4_13 13 0.62 0.33 -0.19 -0.14 0.47 0.53
Q4_14 14 0.53 0.14 0.11 -0.02 0.50 0.50
Q4_15 15 0.80 0.03 -0.09 0.03 0.62 0.38
Q4_16 16 0.66 0.09 -0.22 0.20 0.52 0.48
Q4_17 17 0.77 0.06 -0.03 -0.14 0.50 0.50
Q4_18 18 1.02 -0.14 -0.12 0.02 0.73 0.27
Q4_19 19 0.79 -0.05 0.09 -0.04 0.62 0.38
Q5_1 20 0.37 -0.01 0.08 0.30 0.44 0.56
Q5_2 21 0.05 0.12 0.04 0.58 0.51 0.49
Q5_3 22 0.19 0.14 -0.14 0.58 0.53 0.47
Q5_4 23 -0.10 -0.14 0.08 0.88 0.64 0.36
Q5_5 24 -0.10 -0.22 0.08 0.88 0.59 0.41
Q5_6 25 0.09 0.02 0.01 0.73 0.65 0.35
Q5_7 26 -0.02 0.40 0.10 0.27 0.44 0.56
Q5_8 27 -0.05 0.13 0.08 0.67 0.60 0.40
Q5_9 28 0.20 0.29 -0.05 0.42 0.58 0.42
Q5_10 29 0.19 0.16 0.03 0.33 0.39 0.61
Q5_11 30 0.26 0.12 0.22 0.16 0.44 0.56
Q5_12 31 0.05 0.15 0.22 0.37 0.47 0.53
Q6_1 32 -0.05 0.90 -0.19 -0.04 0.55 0.45
Q6_2 33 -0.18 1.00 -0.15 0.03 0.66 0.34
Q6_3 34 -0.20 0.97 -0.15 0.05 0.62 0.38
Q6_4 35 -0.15 0.92 -0.18 0.02 0.54 0.46
Q6_5 36 0.07 0.37 0.18 -0.07 0.26 0.74
Q6_6 37 0.04 0.85 -0.04 -0.11 0.64 0.36
Q6_7 38 0.11 0.83 0.01 -0.17 0.68 0.32
Q6_8 39 -0.01 0.92 0.01 -0.20 0.68 0.32
Q6_9 40 0.13 0.22 0.47 -0.26 0.35 0.65
Q6_10 41 0.08 0.27 0.35 -0.02 0.37 0.63
Q6_11 42 0.01 0.36 0.38 0.09 0.55 0.45
Q7_1 43 0.15 0.32 0.25 0.16 0.59 0.41
Q7_2 44 0.06 0.15 0.46 0.14 0.51 0.49
Q7_3 45 -0.19 -0.10 0.85 0.12 0.58 0.42
Q7_4 46 -0.02 0.14 0.57 0.05 0.46 0.54
Q7_5 47 0.16 0.04 0.70 -0.08 0.63 0.37
Q7_6 48 0.08 0.11 0.64 -0.05 0.55 0.45
Q7_7 49 -0.02 -0.28 0.88 0.11 0.63 0.37
Q7_8 50 0.15 -0.04 0.65 -0.05 0.50 0.50
Q7_9 51 0.33 0.18 0.23 0.06 0.50 0.50
Q7_10 52 0.40 0.28 0.11 0.00 0.52 0.48
Q7_11 53 0.26 -0.02 0.52 0.00 0.50 0.50
Q7_12 54 -0.01 -0.05 0.72 -0.03 0.44 0.56
Q7_13 55 -0.15 -0.12 0.74 -0.14 0.27 0.73
Q7_14 56 -0.19 -0.22 0.76 0.18 0.44 0.56
Q7_15 57 0.31 0.20 0.19 0.04 0.44 0.56
PA1 PA3 PA2 PA4
SS loadings 10.68 6.66 7.10 5.32
Proportion Var 0.19 0.12 0.12 0.09
Cumulative Var 0.19 0.30 0.43 0.52
Cum. factor Var 0.36 0.58 0.82 1.00
With factor correlations of
PA1 PA3 PA2 PA4
PA1 1.00 0.73 0.70 0.65
PA3 0.73 1.00 0.63 0.58
PA2 0.70 0.63 1.00 0.65
PA4 0.65 0.58 0.65 1.00
Mean item complexity = 1.5
Test of the hypothesis that 4 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1374 and the objective function was 10.73
The root mean square of the residuals (RMSR) is 0.04
The df corrected root mean square of the residuals is 0.04
Fit based upon off diagonal values = 0.99
Measures of factor score adequacy
PA1 PA3 PA2 PA4
Correlation of (regression) scores with factors 0.98 0.98 0.97 0.96
Multiple R square of scores with factors 0.97 0.95 0.94 0.92
Minimum correlation of possible factor scores 0.94 0.91 0.87 0.84print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA3 PA2 PA4
Q4_1 0.806
Q4_2 0.794
Q4_3 0.769
Q4_4 0.645
Q4_5 0.602
Q4_6
Q4_7
Q4_8 0.885
Q4_9 0.783
Q4_10 0.560
Q4_11 0.727
Q4_12 0.522 0.371
Q4_13 0.543
Q4_14 0.478
Q4_15 0.732
Q4_16 0.621
Q4_17 0.741
Q4_18 0.838
Q4_19 0.762
Q5_1 0.355
Q5_2 0.597
Q5_3 0.610
Q5_4 0.970
Q5_5 0.912
Q5_6 0.664
Q5_7 0.376
Q5_8 0.710
Q5_9 0.423
Q5_10
Q5_11
Q5_12 0.351
Q6_1 0.763
Q6_2 0.908
Q6_3 0.897
Q6_4 0.851
Q6_5 0.382
Q6_6 0.700
Q6_7 0.725
Q6_8 0.835
Q6_9 0.578 -0.312
Q6_10 0.317
Q6_11 0.353 0.369
Q7_1
Q7_2 0.394
Q7_3 0.865
Q7_4 0.558
Q7_5 0.660
Q7_6 0.591
Q7_7 0.896
Q7_8 0.571
Q7_9 0.323
Q7_10 0.383
Q7_11 0.434
Q7_12 0.734
Q7_13 0.732
Q7_14 0.751
Q7_15
PA1 PA3 PA2 PA4
SS loadings 9.520 6.311 6.238 4.572
Proportion Var 0.167 0.111 0.109 0.080
Cumulative Var 0.167 0.278 0.387 0.467# rerun the above using the correlation matrix (pearson) to more easily get the standardized factor loadings.
fit <- psych::fa(r = corr.ppm, nfactors = 4, rotate = "Promax", fm = "pa", covar = T)
out.dat[,c(5:8)] <- fit$loadings
out.dat[,9] <- fit$communality
out.dat[,10] <- fit$uniquenesses
write.csv(out.dat, file="data/efa_results_2021_01_06.csv")Five factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 5, rotate = "Promax", fm = "pa")
fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 5, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA1 PA3 PA2 PA4 PA5 h2 u2 com
Q4_1 1.02 -0.17 -0.13 0.01 -0.07 0.67 0.33 1.1
Q4_2 0.99 -0.16 -0.08 0.04 -0.01 0.72 0.28 1.1
Q4_3 1.01 -0.21 -0.05 -0.01 -0.08 0.67 0.33 1.1
Q4_4 0.94 -0.10 0.05 -0.03 -0.25 0.77 0.23 1.2
Q4_5 0.67 0.09 -0.10 0.14 0.02 0.59 0.41 1.2
Q4_6 0.30 0.10 0.11 0.29 -0.12 0.49 0.51 2.8
Q4_7 0.06 0.06 0.29 0.27 -0.16 0.36 0.64 2.7
Q4_8 0.97 -0.02 -0.13 -0.09 0.07 0.68 0.32 1.1
Q4_9 0.74 -0.09 0.14 -0.12 0.10 0.53 0.47 1.2
Q4_10 0.79 -0.04 0.11 -0.05 -0.13 0.63 0.37 1.1
Q4_11 0.72 -0.01 0.15 -0.04 0.10 0.67 0.33 1.1
Q4_12 0.39 0.34 0.09 -0.09 0.28 0.63 0.37 3.1
Q4_13 0.53 0.31 -0.10 -0.07 0.38 0.70 0.30 2.6
Q4_14 0.42 0.12 0.17 0.05 0.28 0.63 0.37 2.3
Q4_15 0.73 0.00 -0.03 0.10 0.18 0.69 0.31 1.2
Q4_16 0.63 0.11 -0.22 0.25 0.08 0.59 0.41 1.7
Q4_17 0.75 0.08 -0.01 -0.11 0.06 0.56 0.44 1.1
Q4_18 1.02 -0.13 -0.11 0.07 0.05 0.83 0.17 1.1
Q4_19 0.71 -0.03 0.15 0.00 0.13 0.68 0.32 1.2
Q5_1 0.32 -0.07 0.12 0.39 0.14 0.52 0.48 2.5
Q5_2 -0.01 0.09 0.02 0.66 0.09 0.56 0.44 1.1
Q5_3 0.09 0.05 -0.11 0.74 0.21 0.66 0.34 1.2
Q5_4 -0.08 -0.21 0.08 0.93 -0.08 0.67 0.33 1.2
Q5_5 -0.05 -0.27 0.06 0.89 -0.13 0.60 0.40 1.2
Q5_6 0.09 -0.04 0.00 0.80 -0.01 0.70 0.30 1.0
Q5_7 -0.09 0.40 0.13 0.32 0.07 0.50 0.50 2.3
Q5_8 -0.09 0.07 0.08 0.76 0.00 0.65 0.35 1.1
Q5_9 0.15 0.26 -0.05 0.53 0.05 0.66 0.34 1.7
Q5_10 0.18 0.18 0.03 0.36 0.01 0.45 0.55 2.1
Q5_11 0.20 0.08 0.27 0.25 0.05 0.51 0.49 3.1
Q5_12 0.01 0.16 0.20 0.43 -0.04 0.51 0.49 1.8
Q6_1 -0.09 0.95 -0.18 -0.01 0.14 0.67 0.33 1.1
Q6_2 -0.21 1.07 -0.18 0.06 -0.02 0.73 0.27 1.1
Q6_3 -0.18 1.05 -0.18 0.06 -0.03 0.72 0.28 1.1
Q6_4 -0.16 1.03 -0.23 0.01 -0.06 0.63 0.37 1.2
Q6_5 0.02 0.40 0.20 -0.03 0.08 0.33 0.67 1.6
Q6_6 0.04 0.96 -0.05 -0.12 -0.07 0.77 0.23 1.1
Q6_7 0.09 0.90 0.01 -0.17 0.03 0.78 0.22 1.1
Q6_8 -0.06 1.01 0.01 -0.19 -0.01 0.77 0.23 1.1
Q6_9 -0.02 0.16 0.64 -0.21 0.30 0.53 0.47 1.8
Q6_10 0.02 0.24 0.40 0.03 0.08 0.41 0.59 1.8
Q6_11 -0.04 0.38 0.39 0.12 0.03 0.59 0.41 2.2
Q7_1 0.13 0.38 0.25 0.17 -0.10 0.67 0.33 2.7
Q7_2 0.08 0.23 0.46 0.10 -0.14 0.58 0.42 1.9
Q7_3 -0.20 -0.08 0.88 0.11 -0.13 0.60 0.40 1.2
Q7_4 0.02 0.28 0.55 -0.03 -0.38 0.63 0.37 2.4
Q7_5 0.17 0.10 0.71 -0.11 -0.15 0.69 0.31 1.3
Q7_6 0.06 0.11 0.67 -0.04 0.00 0.59 0.41 1.1
Q7_7 -0.03 -0.36 0.98 0.14 -0.01 0.72 0.28 1.3
Q7_8 0.15 -0.01 0.68 -0.06 -0.12 0.55 0.45 1.2
Q7_9 0.34 0.26 0.22 0.06 -0.16 0.58 0.42 3.3
Q7_10 0.45 0.37 0.10 -0.01 -0.23 0.66 0.34 2.6
Q7_11 0.33 0.07 0.52 -0.08 -0.23 0.62 0.38 2.3
Q7_12 -0.07 -0.14 0.85 -0.02 0.11 0.53 0.47 1.1
Q7_13 -0.24 -0.26 0.90 -0.10 0.13 0.38 0.62 1.4
Q7_14 -0.18 -0.31 0.84 0.18 -0.07 0.48 0.52 1.5
Q7_15 0.33 0.29 0.17 0.03 -0.20 0.54 0.46 3.2
PA1 PA3 PA2 PA4 PA5
SS loadings 11.88 8.37 7.60 5.75 1.18
Proportion Var 0.21 0.15 0.13 0.10 0.02
Cumulative Var 0.21 0.36 0.49 0.59 0.61
Proportion Explained 0.34 0.24 0.22 0.17 0.03
Cumulative Proportion 0.34 0.58 0.80 0.97 1.00
With factor correlations of
PA1 PA3 PA2 PA4 PA5
PA1 1.00 0.74 0.71 0.68 0.17
PA3 0.74 1.00 0.66 0.64 0.22
PA2 0.71 0.66 1.00 0.69 0.12
PA4 0.68 0.64 0.69 1.00 0.09
PA5 0.17 0.22 0.12 0.09 1.00
Mean item complexity = 1.6
Test of the hypothesis that 5 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1321 and the objective function was 40.74
The root mean square of the residuals (RMSR) is 0.04
The df corrected root mean square of the residuals is 0.04
Fit based upon off diagonal values = 0.99print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA3 PA2 PA4 PA5
Q4_1 1.017
Q4_2 0.991
Q4_3 1.007
Q4_4 0.936
Q4_5 0.666
Q4_6
Q4_7
Q4_8 0.965
Q4_9 0.739
Q4_10 0.787
Q4_11 0.716
Q4_12 0.390 0.338
Q4_13 0.533 0.308 0.382
Q4_14 0.424
Q4_15 0.729
Q4_16 0.625
Q4_17 0.751
Q4_18 1.024
Q4_19 0.710
Q5_1 0.321 0.388
Q5_2 0.662
Q5_3 0.740
Q5_4 0.930
Q5_5 0.895
Q5_6 0.803
Q5_7 0.404 0.317
Q5_8 0.764
Q5_9 0.528
Q5_10 0.358
Q5_11
Q5_12 0.430
Q6_1 0.950
Q6_2 1.066
Q6_3 1.049
Q6_4 1.033
Q6_5 0.396
Q6_6 0.964
Q6_7 0.898
Q6_8 1.015
Q6_9 0.643
Q6_10 0.396
Q6_11 0.384 0.387
Q7_1 0.382
Q7_2 0.457
Q7_3 0.876
Q7_4 0.550 -0.384
Q7_5 0.710
Q7_6 0.669
Q7_7 -0.363 0.977
Q7_8 0.682
Q7_9 0.335
Q7_10 0.445 0.374
Q7_11 0.332 0.519
Q7_12 0.851
Q7_13 0.897
Q7_14 -0.306 0.836
Q7_15 0.334
PA1 PA3 PA2 PA4 PA5
SS loadings 11.823 9.105 7.726 5.384 1.195
Proportion Var 0.207 0.160 0.136 0.094 0.021
Cumulative Var 0.207 0.367 0.503 0.597 0.618fit <- psych::fa(r = cov.dat, nfactors = 5, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 5, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA3 PA4 PA2 PA5 h2 u2 H2 U2
Q4_1 0.82 -0.09 0.01 -0.04 -0.16 0.44 0.31 0.59 0.41
Q4_2 0.79 -0.11 0.01 -0.04 -0.07 0.43 0.24 0.64 0.36
Q4_3 0.79 -0.12 -0.01 0.02 -0.14 0.42 0.28 0.60 0.40
Q4_4 0.70 -0.02 -0.02 0.17 -0.24 0.45 0.22 0.67 0.33
Q4_5 0.58 0.05 0.09 -0.07 -0.01 0.41 0.37 0.52 0.48
Q4_6 0.29 0.14 0.25 0.15 -0.12 0.42 0.54 0.44 0.56
Q4_7 0.10 0.08 0.23 0.31 -0.13 0.30 0.58 0.35 0.65
Q4_8 0.86 -0.01 -0.09 -0.11 -0.02 0.52 0.34 0.61 0.39
Q4_9 0.75 -0.10 -0.14 0.07 0.09 0.48 0.53 0.47 0.53
Q4_10 0.60 0.00 -0.05 0.15 -0.15 0.34 0.28 0.55 0.45
Q4_11 0.69 -0.04 -0.06 0.07 0.12 0.55 0.36 0.60 0.40
Q4_12 0.46 0.23 -0.12 -0.05 0.30 0.57 0.54 0.51 0.49
Q4_13 0.47 0.14 -0.08 -0.22 0.27 0.39 0.38 0.51 0.49
Q4_14 0.41 -0.01 0.03 0.02 0.30 0.43 0.37 0.54 0.46
Q4_15 0.68 -0.05 0.06 -0.10 0.15 0.53 0.31 0.63 0.37
Q4_16 0.60 0.07 0.21 -0.18 -0.04 0.46 0.42 0.52 0.48
Q4_17 0.72 0.06 -0.14 -0.02 0.03 0.46 0.47 0.50 0.50
Q4_18 0.82 -0.10 0.02 -0.08 -0.02 0.50 0.18 0.73 0.27
Q4_19 0.72 -0.08 -0.02 0.05 0.12 0.58 0.36 0.62 0.38
Q5_1 0.28 -0.15 0.36 0.00 0.24 0.44 0.46 0.49 0.51
Q5_2 0.01 0.05 0.67 0.01 0.05 0.56 0.51 0.52 0.48
Q5_3 0.10 -0.07 0.76 -0.22 0.24 0.68 0.41 0.62 0.38
Q5_4 -0.08 -0.09 1.01 0.14 -0.27 0.78 0.45 0.63 0.37
Q5_5 -0.07 -0.13 0.94 0.14 -0.31 0.63 0.44 0.59 0.41
Q5_6 0.06 -0.02 0.72 0.01 -0.05 0.54 0.28 0.66 0.34
Q5_7 -0.05 0.27 0.30 0.06 0.15 0.40 0.49 0.45 0.55
Q5_8 -0.08 0.07 0.78 0.07 0.01 0.67 0.44 0.60 0.40
Q5_9 0.14 0.17 0.50 -0.07 0.13 0.60 0.39 0.60 0.40
Q5_10 0.14 0.10 0.31 0.02 0.03 0.28 0.43 0.39 0.61
Q5_11 0.21 0.03 0.20 0.15 0.18 0.41 0.50 0.45 0.55
Q5_12 0.04 0.12 0.38 0.19 0.02 0.42 0.48 0.47 0.53
Q6_1 -0.06 0.68 -0.02 -0.15 0.08 0.39 0.32 0.55 0.45
Q6_2 -0.13 0.93 0.02 -0.07 -0.12 0.58 0.25 0.69 0.31
Q6_3 -0.15 0.92 0.04 -0.08 -0.12 0.56 0.30 0.65 0.35
Q6_4 -0.10 0.92 0.00 -0.09 -0.18 0.51 0.35 0.59 0.41
Q6_5 0.02 0.25 -0.04 0.12 0.28 0.30 0.77 0.28 0.72
Q6_6 0.04 0.68 -0.09 -0.01 0.02 0.43 0.24 0.64 0.36
Q6_7 0.10 0.67 -0.15 0.01 0.10 0.52 0.24 0.68 0.32
Q6_8 0.00 0.78 -0.18 0.01 0.10 0.56 0.26 0.68 0.32
Q6_9 0.01 -0.05 -0.28 0.38 0.84 0.74 0.75 0.50 0.50
Q6_10 0.02 0.11 0.02 0.22 0.33 0.33 0.50 0.40 0.60
Q6_11 -0.02 0.26 0.11 0.29 0.23 0.52 0.42 0.55 0.45
Q7_1 0.13 0.26 0.16 0.20 0.08 0.48 0.34 0.59 0.41
Q7_2 0.07 0.16 0.11 0.37 0.01 0.38 0.35 0.52 0.48
Q7_3 -0.15 -0.01 0.09 0.80 0.02 0.61 0.42 0.59 0.41
Q7_4 0.06 0.30 -0.02 0.58 -0.18 0.52 0.44 0.54 0.46
Q7_5 0.18 0.08 -0.10 0.59 0.10 0.57 0.33 0.63 0.37
Q7_6 0.08 0.09 -0.05 0.51 0.17 0.46 0.38 0.55 0.45
Q7_7 0.00 -0.23 0.10 0.80 0.11 0.65 0.38 0.63 0.37
Q7_8 0.14 -0.01 -0.05 0.50 0.11 0.39 0.38 0.50 0.50
Q7_9 0.33 0.19 0.06 0.21 0.03 0.49 0.49 0.50 0.50
Q7_10 0.40 0.31 -0.02 0.12 -0.05 0.48 0.41 0.54 0.46
Q7_11 0.25 0.06 -0.03 0.42 -0.04 0.37 0.34 0.52 0.48
Q7_12 -0.04 -0.13 -0.01 0.60 0.33 0.47 0.58 0.45 0.55
Q7_13 -0.20 -0.24 -0.12 0.58 0.42 0.30 0.68 0.31 0.69
Q7_14 -0.16 -0.17 0.18 0.67 0.07 0.43 0.55 0.44 0.56
Q7_15 0.29 0.20 0.04 0.16 0.01 0.36 0.46 0.44 0.56
PA1 PA3 PA4 PA2 PA5
SS loadings 9.10 5.24 5.22 5.46 2.46
Proportion Var 0.18 0.10 0.10 0.11 0.05
Cumulative Var 0.18 0.28 0.39 0.49 0.54
Proportion Explained 0.33 0.19 0.19 0.20 0.09
Cumulative Proportion 0.33 0.52 0.71 0.91 1.00
Standardized loadings (pattern matrix)
item PA1 PA3 PA4 PA2 PA5 h2 u2
Q4_1 1 0.95 -0.11 0.01 -0.04 -0.18 0.59 0.41
Q4_2 2 0.96 -0.13 0.01 -0.05 -0.08 0.64 0.36
Q4_3 3 0.94 -0.14 -0.01 0.03 -0.17 0.60 0.40
Q4_4 4 0.86 -0.02 -0.02 0.20 -0.29 0.67 0.33
Q4_5 5 0.66 0.06 0.10 -0.08 -0.01 0.52 0.48
Q4_6 6 0.30 0.14 0.26 0.16 -0.12 0.44 0.56
Q4_7 7 0.11 0.09 0.25 0.33 -0.14 0.35 0.65
Q4_8 8 0.93 -0.01 -0.10 -0.12 -0.02 0.61 0.39
Q4_9 9 0.75 -0.10 -0.14 0.07 0.09 0.47 0.53
Q4_10 10 0.75 0.00 -0.06 0.19 -0.19 0.55 0.45
Q4_11 11 0.73 -0.04 -0.06 0.07 0.12 0.60 0.40
Q4_12 12 0.43 0.22 -0.12 -0.05 0.28 0.51 0.49
Q4_13 13 0.54 0.15 -0.09 -0.25 0.31 0.51 0.49
Q4_14 14 0.46 -0.02 0.03 0.02 0.33 0.54 0.46
Q4_15 15 0.74 -0.06 0.07 -0.11 0.16 0.63 0.37
Q4_16 16 0.63 0.08 0.22 -0.19 -0.05 0.52 0.48
Q4_17 17 0.75 0.06 -0.15 -0.02 0.03 0.50 0.50
Q4_18 18 0.99 -0.12 0.03 -0.10 -0.03 0.73 0.27
Q4_19 19 0.74 -0.09 -0.02 0.05 0.13 0.62 0.38
Q5_1 20 0.30 -0.16 0.38 0.00 0.26 0.49 0.51
Q5_2 21 0.01 0.05 0.65 0.01 0.04 0.52 0.48
Q5_3 22 0.10 -0.06 0.72 -0.21 0.23 0.62 0.38
Q5_4 23 -0.07 -0.08 0.91 0.13 -0.24 0.63 0.37
Q5_5 24 -0.07 -0.13 0.91 0.13 -0.30 0.59 0.41
Q5_6 25 0.07 -0.02 0.80 0.02 -0.06 0.66 0.34
Q5_7 26 -0.05 0.29 0.32 0.06 0.16 0.45 0.55
Q5_8 27 -0.07 0.06 0.74 0.06 0.01 0.60 0.40
Q5_9 28 0.14 0.17 0.50 -0.07 0.13 0.60 0.40
Q5_10 29 0.16 0.11 0.37 0.02 0.04 0.39 0.61
Q5_11 30 0.22 0.03 0.21 0.16 0.19 0.45 0.55
Q5_12 31 0.04 0.12 0.40 0.20 0.03 0.47 0.53
Q6_1 32 -0.07 0.81 -0.02 -0.18 0.09 0.55 0.45
Q6_2 33 -0.15 1.03 0.02 -0.08 -0.13 0.69 0.31
Q6_3 34 -0.16 1.00 0.04 -0.08 -0.13 0.65 0.35
Q6_4 35 -0.11 0.99 0.00 -0.10 -0.20 0.59 0.41
Q6_5 36 0.02 0.25 -0.04 0.11 0.27 0.28 0.72
Q6_6 37 0.05 0.83 -0.11 -0.02 0.02 0.64 0.36
Q6_7 38 0.11 0.77 -0.17 0.01 0.11 0.68 0.32
Q6_8 39 0.00 0.86 -0.20 0.01 0.11 0.68 0.32
Q6_9 40 0.01 -0.04 -0.23 0.31 0.68 0.50 0.50
Q6_10 41 0.02 0.12 0.02 0.24 0.36 0.40 0.60
Q6_11 42 -0.02 0.27 0.12 0.30 0.24 0.55 0.45
Q7_1 43 0.14 0.29 0.18 0.22 0.09 0.59 0.41
Q7_2 44 0.09 0.19 0.13 0.43 0.01 0.52 0.48
Q7_3 45 -0.15 -0.01 0.09 0.79 0.02 0.59 0.41
Q7_4 46 0.06 0.31 -0.02 0.60 -0.18 0.54 0.46
Q7_5 47 0.18 0.08 -0.10 0.63 0.10 0.63 0.37
Q7_6 48 0.08 0.09 -0.06 0.55 0.18 0.55 0.45
Q7_7 49 0.00 -0.22 0.10 0.78 0.11 0.63 0.37
Q7_8 50 0.16 -0.01 -0.06 0.57 0.13 0.50 0.50
Q7_9 51 0.33 0.19 0.06 0.22 0.03 0.50 0.50
Q7_10 52 0.42 0.33 -0.02 0.13 -0.05 0.54 0.46
Q7_11 53 0.30 0.07 -0.03 0.50 -0.04 0.52 0.48
Q7_12 54 -0.04 -0.13 -0.01 0.58 0.33 0.45 0.55
Q7_13 55 -0.20 -0.24 -0.12 0.58 0.43 0.31 0.69
Q7_14 56 -0.16 -0.18 0.18 0.68 0.07 0.44 0.56
Q7_15 57 0.32 0.22 0.04 0.18 0.01 0.44 0.56
PA1 PA3 PA4 PA2 PA5
SS loadings 10.22 5.88 5.86 6.14 2.76
Proportion Var 0.18 0.10 0.10 0.11 0.05
Cumulative Var 0.18 0.28 0.39 0.49 0.54
Cum. factor Var 0.33 0.52 0.71 0.91 1.00
With factor correlations of
PA1 PA3 PA4 PA2 PA5
PA1 1.00 0.70 0.69 0.61 0.62
PA3 0.70 1.00 0.62 0.51 0.65
PA4 0.69 0.62 1.00 0.63 0.58
PA2 0.61 0.51 0.63 1.00 0.42
PA5 0.62 0.65 0.58 0.42 1.00
Mean item complexity = 1.7
Test of the hypothesis that 5 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1321 and the objective function was 9.85
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.04
Fit based upon off diagonal values = 0.99
Measures of factor score adequacy
PA1 PA3 PA4 PA2 PA5
Correlation of (regression) scores with factors 0.98 0.98 0.97 0.96 0.92
Multiple R square of scores with factors 0.97 0.95 0.93 0.92 0.84
Minimum correlation of possible factor scores 0.94 0.90 0.87 0.84 0.68print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA3 PA4 PA2 PA5
Q4_1 0.820
Q4_2 0.785
Q4_3 0.785
Q4_4 0.701
Q4_5 0.583
Q4_6
Q4_7 0.311
Q4_8 0.865
Q4_9 0.754
Q4_10 0.595
Q4_11 0.692
Q4_12 0.456
Q4_13 0.470
Q4_14 0.409
Q4_15 0.676
Q4_16 0.597
Q4_17 0.723
Q4_18 0.815
Q4_19 0.721
Q5_1 0.360
Q5_2 0.672
Q5_3 0.757
Q5_4 1.009
Q5_5 0.941 -0.310
Q5_6 0.722
Q5_7 0.302
Q5_8 0.783
Q5_9 0.496
Q5_10 0.312
Q5_11
Q5_12 0.383
Q6_1 0.682
Q6_2 0.935
Q6_3 0.924
Q6_4 0.917
Q6_5
Q6_6 0.679
Q6_7 0.671
Q6_8 0.777
Q6_9 0.383 0.836
Q6_10 0.328
Q6_11
Q7_1
Q7_2 0.368
Q7_3 0.802
Q7_4 0.584
Q7_5 0.595
Q7_6 0.509
Q7_7 0.796
Q7_8 0.504
Q7_9 0.326
Q7_10 0.401 0.310
Q7_11 0.418
Q7_12 0.596 0.333
Q7_13 0.578 0.422
Q7_14 0.670
Q7_15
PA1 PA3 PA4 PA2 PA5
SS loadings 8.951 5.582 5.349 4.857 2.260
Proportion Var 0.157 0.098 0.094 0.085 0.040
Cumulative Var 0.157 0.255 0.349 0.434 0.474Six factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 6,
rotate = "Promax", fm = "pa")
fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 6, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA1 PA6 PA2 PA4 PA5 PA3 h2 u2 com
Q4_1 0.85 -0.12 -0.10 0.01 0.18 -0.07 0.67 0.33 1.2
Q4_2 0.82 -0.14 -0.08 0.03 0.15 0.05 0.72 0.28 1.2
Q4_3 0.86 -0.13 -0.02 0.00 0.14 -0.08 0.68 0.32 1.1
Q4_4 0.83 0.00 0.11 -0.02 0.21 -0.30 0.78 0.22 1.4
Q4_5 0.52 0.03 -0.12 0.13 0.18 0.10 0.59 0.41 1.6
Q4_6 0.30 0.15 0.16 0.29 0.06 -0.15 0.50 0.50 3.7
Q4_7 0.15 0.20 0.38 0.28 -0.09 -0.23 0.42 0.58 3.8
Q4_8 0.86 0.04 -0.05 -0.08 -0.06 0.11 0.69 0.31 1.1
Q4_9 0.65 -0.04 0.14 -0.11 -0.06 0.18 0.53 0.47 1.4
Q4_10 0.77 0.11 0.21 -0.04 -0.03 -0.18 0.68 0.32 1.3
Q4_11 0.63 0.02 0.14 -0.03 -0.02 0.20 0.67 0.33 1.3
Q4_12 0.39 0.33 0.13 -0.08 -0.28 0.43 0.65 0.35 4.0
Q4_13 0.42 0.18 -0.14 -0.07 -0.13 0.60 0.71 0.29 2.3
Q4_14 0.37 0.09 0.13 0.05 -0.16 0.45 0.62 0.38 2.5
Q4_15 0.62 0.00 -0.02 0.10 -0.06 0.29 0.69 0.31 1.5
Q4_16 0.57 0.15 -0.13 0.25 -0.09 0.09 0.60 0.40 1.8
Q4_17 0.67 0.10 0.03 -0.10 -0.02 0.12 0.56 0.44 1.2
Q4_18 0.87 -0.09 -0.08 0.07 0.04 0.11 0.83 0.17 1.1
Q4_19 0.63 0.01 0.15 0.00 -0.08 0.24 0.69 0.31 1.4
Q5_1 0.16 -0.21 -0.04 0.37 0.22 0.33 0.57 0.43 3.8
Q5_2 -0.01 0.07 0.03 0.64 -0.05 0.15 0.56 0.44 1.2
Q5_3 -0.02 -0.08 -0.19 0.72 0.05 0.36 0.69 0.31 1.7
Q5_4 -0.01 -0.07 0.18 0.91 -0.13 -0.16 0.69 0.31 1.2
Q5_5 0.01 -0.12 0.15 0.87 -0.08 -0.22 0.61 0.39 1.3
Q5_6 0.07 -0.02 0.02 0.77 0.02 0.01 0.70 0.30 1.0
Q5_7 -0.04 0.36 0.14 0.31 -0.07 0.16 0.50 0.50 2.9
Q5_8 -0.09 0.04 0.07 0.73 0.06 0.05 0.65 0.35 1.1
Q5_9 0.05 0.11 -0.13 0.50 0.23 0.17 0.68 0.32 1.9
Q5_10 0.12 0.11 -0.01 0.34 0.13 0.10 0.45 0.55 2.0
Q5_11 0.08 -0.05 0.10 0.24 0.28 0.22 0.53 0.47 3.5
Q5_12 -0.03 0.08 0.12 0.41 0.20 0.04 0.51 0.49 1.8
Q6_1 -0.08 0.74 -0.17 -0.01 0.05 0.24 0.67 0.33 1.4
Q6_2 -0.10 0.96 -0.06 0.07 -0.03 -0.04 0.76 0.24 1.0
Q6_3 -0.04 1.00 -0.02 0.08 -0.13 -0.07 0.79 0.21 1.1
Q6_4 -0.04 0.97 -0.07 0.02 -0.08 -0.11 0.68 0.32 1.1
Q6_5 -0.06 0.20 0.05 -0.04 0.26 0.26 0.35 0.65 3.1
Q6_6 0.02 0.73 -0.08 -0.11 0.29 0.04 0.76 0.24 1.4
Q6_7 0.11 0.75 0.03 -0.16 0.06 0.13 0.78 0.22 1.2
Q6_8 -0.04 0.80 -0.01 -0.18 0.18 0.11 0.76 0.24 1.2
Q6_9 -0.05 0.04 0.43 -0.20 -0.02 0.58 0.53 0.47 2.2
Q6_10 -0.06 0.09 0.22 0.02 0.23 0.27 0.42 0.58 3.2
Q6_11 -0.02 0.30 0.31 0.12 0.08 0.16 0.59 0.41 3.0
Q7_1 -0.04 0.12 0.01 0.15 0.63 0.08 0.73 0.27 1.2
Q7_2 0.04 0.15 0.33 0.09 0.31 -0.04 0.57 0.43 2.6
Q7_3 -0.05 0.11 0.89 0.12 -0.18 -0.12 0.68 0.32 1.2
Q7_4 -0.02 0.19 0.40 -0.03 0.54 -0.33 0.62 0.38 2.9
Q7_5 0.10 0.02 0.51 -0.11 0.38 0.01 0.69 0.31 2.1
Q7_6 0.09 0.13 0.57 -0.03 0.01 0.13 0.60 0.40 1.3
Q7_7 0.08 -0.16 0.93 0.14 -0.21 0.06 0.78 0.22 1.2
Q7_8 0.08 -0.08 0.48 -0.06 0.33 0.04 0.55 0.45 1.9
Q7_9 0.09 -0.04 -0.07 0.02 0.82 0.02 0.70 0.30 1.0
Q7_10 0.19 0.08 -0.16 -0.04 0.88 -0.10 0.77 0.23 1.2
Q7_11 0.18 -0.06 0.29 -0.09 0.59 -0.10 0.64 0.36 1.9
Q7_12 -0.10 -0.18 0.61 -0.02 0.11 0.32 0.53 0.47 1.9
Q7_13 -0.19 -0.21 0.71 -0.10 -0.09 0.29 0.37 0.63 1.8
Q7_14 -0.08 -0.14 0.78 0.18 -0.12 -0.05 0.50 0.50 1.3
Q7_15 0.10 0.01 -0.09 0.00 0.82 -0.06 0.64 0.36 1.1
PA1 PA6 PA2 PA4 PA5 PA3
SS loadings 9.63 6.37 5.74 5.54 5.12 3.52
Proportion Var 0.17 0.11 0.10 0.10 0.09 0.06
Cumulative Var 0.17 0.28 0.38 0.48 0.57 0.63
Proportion Explained 0.27 0.18 0.16 0.15 0.14 0.10
Cumulative Proportion 0.27 0.45 0.61 0.76 0.90 1.00
With factor correlations of
PA1 PA6 PA2 PA4 PA5 PA3
PA1 1.00 0.60 0.56 0.63 0.69 0.59
PA6 0.60 1.00 0.46 0.55 0.66 0.61
PA2 0.56 0.46 1.00 0.60 0.68 0.46
PA4 0.63 0.55 0.60 1.00 0.66 0.52
PA5 0.69 0.66 0.68 0.66 1.00 0.66
PA3 0.59 0.61 0.46 0.52 0.66 1.00
Mean item complexity = 1.8
Test of the hypothesis that 6 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1269 and the objective function was 39.16
The root mean square of the residuals (RMSR) is 0.04
The df corrected root mean square of the residuals is 0.04
Fit based upon off diagonal values = 0.99print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA6 PA2 PA4 PA5 PA3
Q4_1 0.849
Q4_2 0.815
Q4_3 0.859
Q4_4 0.835
Q4_5 0.523
Q4_6
Q4_7 0.377
Q4_8 0.857
Q4_9 0.653
Q4_10 0.767
Q4_11 0.626
Q4_12 0.392 0.325 0.428
Q4_13 0.420 0.598
Q4_14 0.369 0.446
Q4_15 0.618
Q4_16 0.567
Q4_17 0.666
Q4_18 0.869
Q4_19 0.628
Q5_1 0.370 0.334
Q5_2 0.637
Q5_3 0.715 0.363
Q5_4 0.910
Q5_5 0.873
Q5_6 0.773
Q5_7 0.355 0.311
Q5_8 0.734
Q5_9 0.505
Q5_10 0.344
Q5_11
Q5_12 0.410
Q6_1 0.738
Q6_2 0.959
Q6_3 1.005
Q6_4 0.974
Q6_5
Q6_6 0.734
Q6_7 0.749
Q6_8 0.799
Q6_9 0.432 0.582
Q6_10
Q6_11 0.304 0.308
Q7_1 0.629
Q7_2 0.331 0.307
Q7_3 0.891
Q7_4 0.399 0.544 -0.326
Q7_5 0.507 0.378
Q7_6 0.573
Q7_7 0.928
Q7_8 0.484 0.328
Q7_9 0.821
Q7_10 0.880
Q7_11 0.589
Q7_12 0.610 0.323
Q7_13 0.707
Q7_14 0.777
Q7_15 0.815
PA1 PA6 PA2 PA4 PA5 PA3
SS loadings 8.393 6.097 5.277 5.044 4.466 2.648
Proportion Var 0.147 0.107 0.093 0.088 0.078 0.046
Cumulative Var 0.147 0.254 0.347 0.435 0.514 0.560fit <- psych::fa(r = cov.dat, nfactors = 6, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 6, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA4 PA3 PA5 PA2 PA6 h2 u2 H2 U2
Q4_1 0.68 0.01 -0.10 0.21 -0.06 -0.17 0.44 0.31 0.59 0.41
Q4_2 0.68 0.01 -0.10 0.12 -0.05 -0.07 0.43 0.24 0.64 0.36
Q4_3 0.67 0.00 -0.10 0.14 0.01 -0.15 0.41 0.28 0.60 0.40
Q4_4 0.60 0.00 -0.01 0.19 0.12 -0.25 0.44 0.22 0.67 0.33
Q4_5 0.48 0.08 0.02 0.17 -0.09 -0.01 0.41 0.37 0.52 0.48
Q4_6 0.26 0.26 0.14 0.10 0.13 -0.12 0.42 0.54 0.44 0.56
Q4_7 0.15 0.26 0.14 -0.09 0.33 -0.13 0.32 0.56 0.37 0.63
Q4_8 0.81 -0.07 0.03 -0.04 -0.05 -0.01 0.53 0.33 0.62 0.38
Q4_9 0.73 -0.11 -0.03 -0.08 0.12 0.10 0.50 0.52 0.49 0.51
Q4_10 0.56 -0.02 0.05 0.01 0.17 -0.15 0.35 0.27 0.56 0.44
Q4_11 0.64 -0.04 -0.01 0.00 0.09 0.14 0.56 0.36 0.61 0.39
Q4_12 0.52 -0.09 0.29 -0.28 0.07 0.36 0.61 0.49 0.55 0.45
Q4_13 0.48 -0.06 0.16 -0.16 -0.12 0.31 0.40 0.37 0.52 0.48
Q4_14 0.42 0.04 0.03 -0.12 0.07 0.33 0.44 0.36 0.55 0.45
Q4_15 0.63 0.07 -0.02 -0.04 -0.05 0.17 0.53 0.31 0.63 0.37
Q4_16 0.56 0.22 0.10 -0.05 -0.10 -0.03 0.47 0.42 0.53 0.47
Q4_17 0.68 -0.12 0.09 -0.01 0.02 0.04 0.47 0.46 0.51 0.49
Q4_18 0.73 0.03 -0.07 0.04 -0.05 -0.02 0.50 0.18 0.73 0.27
Q4_19 0.69 0.00 -0.02 -0.07 0.10 0.14 0.60 0.34 0.63 0.37
Q5_1 0.18 0.32 -0.21 0.21 -0.07 0.26 0.46 0.45 0.51 0.49
Q5_2 0.03 0.66 0.06 -0.04 0.04 0.06 0.56 0.51 0.52 0.48
Q5_3 0.06 0.72 -0.09 0.04 -0.21 0.26 0.68 0.41 0.62 0.38
Q5_4 -0.02 1.04 0.00 -0.17 0.20 -0.27 0.80 0.42 0.65 0.35
Q5_5 -0.03 0.96 -0.06 -0.11 0.18 -0.32 0.64 0.43 0.60 0.40
Q5_6 0.05 0.71 -0.01 0.03 0.02 -0.05 0.54 0.28 0.65 0.35
Q5_7 -0.01 0.31 0.25 -0.05 0.08 0.18 0.40 0.48 0.45 0.55
Q5_8 -0.09 0.76 0.04 0.09 0.04 0.02 0.67 0.44 0.60 0.40
Q5_9 0.05 0.46 0.06 0.28 -0.15 0.14 0.62 0.37 0.62 0.38
Q5_10 0.10 0.30 0.06 0.11 -0.01 0.04 0.28 0.42 0.39 0.61
Q5_11 0.11 0.16 -0.05 0.30 0.03 0.19 0.43 0.48 0.47 0.53
Q5_12 0.00 0.36 0.06 0.20 0.11 0.03 0.43 0.48 0.47 0.53
Q6_1 -0.02 -0.01 0.60 0.00 -0.11 0.10 0.39 0.32 0.55 0.45
Q6_2 -0.06 0.05 0.86 -0.01 -0.01 -0.11 0.59 0.24 0.71 0.29
Q6_3 -0.05 0.08 0.88 -0.11 0.02 -0.11 0.59 0.26 0.70 0.30
Q6_4 -0.02 0.03 0.86 -0.05 -0.01 -0.18 0.54 0.32 0.62 0.38
Q6_5 -0.03 -0.06 0.15 0.24 0.02 0.30 0.31 0.76 0.29 0.71
Q6_6 0.02 -0.09 0.56 0.19 -0.04 0.02 0.43 0.24 0.64 0.36
Q6_7 0.11 -0.14 0.59 0.07 0.02 0.12 0.52 0.24 0.68 0.32
Q6_8 -0.01 -0.18 0.64 0.18 -0.02 0.12 0.55 0.26 0.68 0.32
Q6_9 0.06 -0.28 -0.04 -0.08 0.33 0.90 0.74 0.75 0.49 0.51
Q6_10 -0.03 0.00 0.03 0.21 0.12 0.35 0.34 0.50 0.41 0.59
Q6_11 -0.01 0.11 0.22 0.08 0.24 0.26 0.52 0.42 0.55 0.45
Q7_1 -0.04 0.10 0.08 0.59 -0.01 0.06 0.54 0.28 0.66 0.34
Q7_2 0.01 0.10 0.09 0.28 0.24 0.01 0.38 0.35 0.52 0.48
Q7_3 -0.03 0.14 0.10 -0.15 0.79 0.04 0.66 0.36 0.65 0.35
Q7_4 -0.07 -0.05 0.16 0.57 0.36 -0.21 0.55 0.41 0.57 0.43
Q7_5 0.08 -0.12 0.00 0.41 0.40 0.10 0.58 0.32 0.64 0.36
Q7_6 0.10 -0.03 0.10 0.04 0.45 0.19 0.47 0.38 0.55 0.45
Q7_7 0.10 0.15 -0.09 -0.19 0.79 0.14 0.70 0.33 0.68 0.32
Q7_8 0.08 -0.06 -0.06 0.28 0.36 0.11 0.39 0.38 0.50 0.50
Q7_9 0.06 -0.03 -0.05 0.86 -0.08 -0.02 0.63 0.35 0.64 0.36
Q7_10 0.14 -0.11 0.07 0.85 -0.14 -0.11 0.61 0.28 0.68 0.32
Q7_11 0.11 -0.07 -0.05 0.50 0.22 -0.06 0.39 0.31 0.56 0.44
Q7_12 -0.05 -0.01 -0.13 0.12 0.47 0.36 0.47 0.58 0.45 0.55
Q7_13 -0.12 -0.10 -0.16 -0.12 0.53 0.46 0.31 0.67 0.31 0.69
Q7_14 -0.07 0.21 -0.07 -0.13 0.64 0.09 0.45 0.53 0.46 0.54
Q7_15 0.06 -0.05 -0.01 0.76 -0.09 -0.04 0.47 0.35 0.57 0.43
PA1 PA4 PA3 PA5 PA2 PA6
SS loadings 7.76 5.12 4.31 4.38 4.13 2.74
Proportion Var 0.15 0.10 0.08 0.09 0.08 0.05
Cumulative Var 0.15 0.25 0.34 0.42 0.51 0.56
Proportion Explained 0.27 0.18 0.15 0.15 0.15 0.10
Cumulative Proportion 0.27 0.45 0.60 0.76 0.90 1.00
Standardized loadings (pattern matrix)
item PA1 PA4 PA3 PA5 PA2 PA6 h2 u2
Q4_1 1 0.79 0.01 -0.12 0.24 -0.07 -0.20 0.59 0.41
Q4_2 2 0.83 0.01 -0.12 0.14 -0.06 -0.09 0.64 0.36
Q4_3 3 0.81 0.00 -0.12 0.17 0.01 -0.19 0.60 0.40
Q4_4 4 0.73 -0.01 -0.02 0.23 0.15 -0.31 0.67 0.33
Q4_5 5 0.55 0.09 0.02 0.19 -0.11 -0.01 0.52 0.48
Q4_6 6 0.27 0.27 0.14 0.10 0.14 -0.12 0.44 0.56
Q4_7 7 0.16 0.28 0.15 -0.09 0.35 -0.13 0.37 0.63
Q4_8 8 0.87 -0.08 0.04 -0.04 -0.05 -0.01 0.62 0.38
Q4_9 9 0.72 -0.11 -0.03 -0.08 0.12 0.10 0.49 0.51
Q4_10 10 0.71 -0.02 0.06 0.01 0.22 -0.19 0.56 0.44
Q4_11 11 0.67 -0.04 -0.01 -0.01 0.09 0.14 0.61 0.39
Q4_12 12 0.50 -0.08 0.28 -0.27 0.06 0.34 0.55 0.45
Q4_13 13 0.54 -0.07 0.18 -0.18 -0.14 0.35 0.52 0.48
Q4_14 14 0.46 0.05 0.03 -0.13 0.07 0.37 0.55 0.45
Q4_15 15 0.68 0.08 -0.02 -0.05 -0.06 0.18 0.63 0.37
Q4_16 16 0.60 0.24 0.11 -0.06 -0.11 -0.04 0.53 0.47
Q4_17 17 0.70 -0.12 0.09 -0.01 0.02 0.05 0.51 0.49
Q4_18 18 0.88 0.04 -0.08 0.05 -0.07 -0.02 0.73 0.27
Q4_19 19 0.71 0.00 -0.02 -0.07 0.10 0.15 0.63 0.37
Q5_1 20 0.19 0.34 -0.22 0.22 -0.08 0.27 0.51 0.49
Q5_2 21 0.03 0.64 0.06 -0.04 0.04 0.06 0.52 0.48
Q5_3 22 0.06 0.69 -0.09 0.04 -0.20 0.24 0.62 0.38
Q5_4 23 -0.02 0.94 0.00 -0.15 0.18 -0.24 0.65 0.35
Q5_5 24 -0.03 0.92 -0.06 -0.11 0.17 -0.31 0.60 0.40
Q5_6 25 0.05 0.78 -0.01 0.03 0.02 -0.05 0.65 0.35
Q5_7 26 -0.01 0.33 0.27 -0.05 0.08 0.20 0.45 0.55
Q5_8 27 -0.09 0.72 0.04 0.08 0.03 0.02 0.60 0.40
Q5_9 28 0.05 0.46 0.06 0.28 -0.15 0.14 0.62 0.38
Q5_10 29 0.12 0.36 0.07 0.13 -0.01 0.05 0.39 0.61
Q5_11 30 0.11 0.17 -0.06 0.32 0.03 0.20 0.47 0.53
Q5_12 31 0.00 0.38 0.07 0.21 0.12 0.03 0.47 0.53
Q6_1 32 -0.02 -0.01 0.71 0.00 -0.13 0.11 0.55 0.45
Q6_2 33 -0.07 0.05 0.94 -0.01 -0.01 -0.12 0.71 0.29
Q6_3 34 -0.05 0.08 0.95 -0.12 0.02 -0.12 0.70 0.30
Q6_4 35 -0.02 0.03 0.93 -0.05 -0.01 -0.19 0.62 0.38
Q6_5 36 -0.03 -0.06 0.14 0.24 0.02 0.29 0.29 0.71
Q6_6 37 0.03 -0.11 0.68 0.23 -0.05 0.03 0.64 0.36
Q6_7 38 0.13 -0.16 0.67 0.08 0.02 0.13 0.68 0.32
Q6_8 39 -0.01 -0.20 0.71 0.20 -0.03 0.13 0.68 0.32
Q6_9 40 0.05 -0.23 -0.03 -0.06 0.27 0.74 0.49 0.51
Q6_10 41 -0.03 0.00 0.03 0.23 0.13 0.38 0.41 0.59
Q6_11 42 -0.01 0.12 0.22 0.09 0.25 0.27 0.55 0.45
Q7_1 43 -0.04 0.11 0.09 0.65 -0.01 0.07 0.66 0.34
Q7_2 44 0.02 0.11 0.11 0.33 0.29 0.01 0.52 0.48
Q7_3 45 -0.03 0.14 0.10 -0.15 0.78 0.04 0.65 0.35
Q7_4 46 -0.07 -0.05 0.16 0.58 0.36 -0.21 0.57 0.43
Q7_5 47 0.09 -0.12 0.00 0.43 0.43 0.10 0.64 0.36
Q7_6 48 0.11 -0.04 0.11 0.04 0.49 0.21 0.55 0.45
Q7_7 49 0.10 0.15 -0.09 -0.19 0.78 0.14 0.68 0.32
Q7_8 50 0.09 -0.07 -0.07 0.32 0.41 0.13 0.50 0.50
Q7_9 51 0.06 -0.03 -0.05 0.87 -0.08 -0.02 0.64 0.36
Q7_10 52 0.15 -0.11 0.08 0.90 -0.15 -0.12 0.68 0.32
Q7_11 53 0.13 -0.08 -0.06 0.60 0.26 -0.07 0.56 0.44
Q7_12 54 -0.04 -0.01 -0.13 0.12 0.46 0.35 0.45 0.55
Q7_13 55 -0.12 -0.11 -0.16 -0.12 0.53 0.47 0.31 0.69
Q7_14 56 -0.07 0.21 -0.07 -0.13 0.65 0.10 0.46 0.54
Q7_15 57 0.07 -0.05 -0.02 0.84 -0.10 -0.04 0.57 0.43
PA1 PA4 PA3 PA5 PA2 PA6
SS loadings 8.71 5.75 4.84 4.92 4.64 3.08
Proportion Var 0.15 0.10 0.08 0.09 0.08 0.05
Cumulative Var 0.15 0.25 0.34 0.42 0.51 0.56
Cum. factor Var 0.27 0.45 0.60 0.76 0.90 1.00
With factor correlations of
PA1 PA4 PA3 PA5 PA2 PA6
PA1 1.00 0.65 0.59 0.73 0.45 0.62
PA4 0.65 1.00 0.55 0.72 0.51 0.61
PA3 0.59 0.55 1.00 0.65 0.31 0.64
PA5 0.73 0.72 0.65 1.00 0.60 0.69
PA2 0.45 0.51 0.31 0.60 1.00 0.35
PA6 0.62 0.61 0.64 0.69 0.35 1.00
Mean item complexity = 1.7
Test of the hypothesis that 6 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1269 and the objective function was 8.78
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.03
Fit based upon off diagonal values = 0.99
Measures of factor score adequacy
PA1 PA4 PA3 PA5 PA2 PA6
Correlation of (regression) scores with factors 0.98 0.97 0.97 0.97 0.95 0.93
Multiple R square of scores with factors 0.96 0.93 0.94 0.94 0.90 0.86
Minimum correlation of possible factor scores 0.92 0.86 0.88 0.87 0.80 0.72print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA4 PA3 PA5 PA2 PA6
Q4_1 0.684
Q4_2 0.678
Q4_3 0.674
Q4_4 0.599
Q4_5 0.484
Q4_6
Q4_7 0.331
Q4_8 0.805
Q4_9 0.726
Q4_10 0.561
Q4_11 0.644
Q4_12 0.523 0.358
Q4_13 0.477 0.308
Q4_14 0.416 0.331
Q4_15 0.628
Q4_16 0.562
Q4_17 0.677
Q4_18 0.727
Q4_19 0.690
Q5_1 0.321
Q5_2 0.664
Q5_3 0.719
Q5_4 1.035
Q5_5 0.956 -0.317
Q5_6 0.708
Q5_7 0.309
Q5_8 0.760
Q5_9 0.456
Q5_10
Q5_11 0.305
Q5_12 0.364
Q6_1 0.601
Q6_2 0.856
Q6_3 0.883
Q6_4 0.859
Q6_5
Q6_6 0.557
Q6_7 0.587
Q6_8 0.643
Q6_9 0.334 0.899
Q6_10 0.347
Q6_11
Q7_1 0.588
Q7_2
Q7_3 0.789
Q7_4 0.572 0.357
Q7_5 0.405 0.403
Q7_6 0.450
Q7_7 0.789
Q7_8 0.357
Q7_9 0.860
Q7_10 0.848
Q7_11 0.504
Q7_12 0.472 0.357
Q7_13 0.528 0.461
Q7_14 0.639
Q7_15 0.757
PA1 PA4 PA3 PA5 PA2 PA6
SS loadings 7.021 5.244 4.229 4.209 3.544 2.665
Proportion Var 0.123 0.092 0.074 0.074 0.062 0.047
Cumulative Var 0.123 0.215 0.289 0.363 0.425 0.472Seven factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 7, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 7, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA3 PA4 PA2 PA5 PA6 PA1 PA7 h2 u2 com
Q4_1 0.88 -0.01 0.01 0.22 -0.16 -0.18 0.08 0.74 0.26 1.3
Q4_2 0.77 0.03 -0.10 0.15 -0.06 0.07 -0.02 0.72 0.28 1.2
Q4_3 0.88 -0.02 0.00 0.17 -0.09 -0.18 0.10 0.74 0.26 1.2
Q4_4 0.79 -0.04 0.01 0.19 0.23 -0.25 -0.08 0.78 0.22 1.6
Q4_5 0.47 0.14 0.00 0.17 -0.06 0.16 -0.08 0.59 0.41 1.8
Q4_6 0.33 0.28 0.22 0.08 0.12 -0.22 0.08 0.53 0.47 4.3
Q4_7 0.14 0.28 0.18 -0.11 0.41 -0.19 0.01 0.42 0.58 3.2
Q4_8 0.79 -0.09 0.02 -0.06 -0.03 0.22 -0.06 0.69 0.31 1.2
Q4_9 0.58 -0.11 -0.06 -0.07 0.12 0.30 0.02 0.53 0.47 1.7
Q4_10 0.71 -0.05 0.09 -0.05 0.28 -0.08 -0.04 0.68 0.32 1.4
Q4_11 0.57 -0.03 0.00 -0.03 0.10 0.29 0.05 0.67 0.33 1.6
Q4_12 0.29 -0.07 0.18 -0.31 0.13 0.70 -0.05 0.69 0.31 2.1
Q4_13 0.34 -0.05 0.09 -0.12 -0.20 0.77 0.00 0.72 0.28 1.6
Q4_14 0.35 0.06 0.11 -0.12 -0.05 0.47 0.19 0.63 0.37 2.6
Q4_15 0.55 0.11 -0.02 -0.05 -0.06 0.38 0.00 0.69 0.31 1.9
Q4_16 0.55 0.25 0.19 -0.07 -0.16 0.10 0.00 0.60 0.40 2.0
Q4_17 0.59 -0.10 0.03 -0.04 0.09 0.28 -0.08 0.57 0.43 1.6
Q4_18 0.78 0.07 -0.13 0.02 0.00 0.24 -0.12 0.84 0.16 1.3
Q4_19 0.55 0.01 -0.05 -0.09 0.14 0.38 -0.01 0.70 0.30 2.0
Q5_1 0.13 0.40 -0.21 0.23 -0.10 0.31 0.04 0.57 0.43 3.7
Q5_2 -0.03 0.66 0.04 -0.05 0.03 0.16 -0.04 0.56 0.44 1.2
Q5_3 -0.03 0.75 -0.08 0.08 -0.24 0.32 -0.01 0.69 0.31 1.7
Q5_4 0.01 0.93 -0.02 -0.12 0.16 -0.24 0.00 0.69 0.31 1.2
Q5_5 0.04 0.89 -0.06 -0.07 0.13 -0.33 0.01 0.62 0.38 1.3
Q5_6 0.05 0.80 -0.03 0.02 0.04 -0.01 -0.06 0.70 0.30 1.0
Q5_7 -0.09 0.34 0.23 -0.09 0.20 0.30 -0.08 0.53 0.47 4.0
Q5_8 -0.12 0.77 0.00 0.05 0.11 0.06 -0.08 0.66 0.34 1.1
Q5_9 0.03 0.53 0.07 0.24 -0.09 0.16 -0.07 0.68 0.32 1.8
Q5_10 0.04 0.38 -0.03 0.10 0.13 0.24 -0.19 0.50 0.50 2.7
Q5_11 0.12 0.25 0.04 0.32 -0.06 0.08 0.23 0.56 0.44 3.3
Q5_12 -0.03 0.43 0.06 0.20 0.14 0.03 0.01 0.51 0.49 1.7
Q6_1 0.00 -0.02 0.82 0.11 -0.33 0.14 0.20 0.77 0.23 1.6
Q6_2 -0.03 0.06 0.99 0.00 -0.10 -0.10 0.08 0.83 0.17 1.1
Q6_3 -0.01 0.07 0.95 -0.12 0.01 -0.03 -0.01 0.81 0.19 1.0
Q6_4 -0.01 0.01 0.91 -0.07 0.00 -0.06 -0.06 0.69 0.31 1.0
Q6_5 -0.03 -0.03 0.22 0.29 -0.06 0.21 0.16 0.36 0.64 3.6
Q6_6 -0.03 -0.11 0.57 0.27 0.09 0.19 -0.17 0.77 0.23 2.0
Q6_7 0.07 -0.16 0.61 0.05 0.12 0.30 -0.08 0.79 0.21 1.8
Q6_8 -0.07 -0.17 0.64 0.17 0.11 0.25 -0.10 0.77 0.23 1.8
Q6_9 -0.07 -0.18 0.01 -0.01 0.21 0.65 0.29 0.53 0.47 1.9
Q6_10 -0.04 0.04 0.09 0.25 0.11 0.24 0.17 0.42 0.58 3.6
Q6_11 -0.05 0.13 0.22 0.07 0.30 0.24 0.05 0.60 0.40 3.6
Q7_1 -0.02 0.16 0.12 0.64 0.03 -0.01 0.06 0.74 0.26 1.2
Q7_2 0.00 0.10 0.05 0.28 0.42 0.05 -0.03 0.59 0.41 1.9
Q7_3 -0.07 0.13 0.04 -0.21 0.85 -0.02 0.15 0.69 0.31 1.3
Q7_4 -0.06 -0.04 0.06 0.50 0.63 -0.25 -0.13 0.66 0.34 2.4
Q7_5 0.06 -0.10 -0.06 0.35 0.55 0.09 0.06 0.70 0.30 1.9
Q7_6 0.06 -0.02 0.07 0.00 0.51 0.22 0.14 0.60 0.40 1.6
Q7_7 0.05 0.15 -0.17 -0.22 0.77 0.11 0.27 0.77 0.23 1.7
Q7_8 0.01 -0.05 -0.20 0.29 0.57 0.17 0.00 0.60 0.40 2.0
Q7_9 0.12 0.03 0.00 0.84 -0.04 -0.12 0.06 0.71 0.29 1.1
Q7_10 0.20 -0.04 0.07 0.87 -0.01 -0.17 -0.07 0.77 0.23 1.2
Q7_11 0.17 -0.09 -0.07 0.57 0.35 -0.12 0.06 0.63 0.37 2.1
Q7_12 -0.02 -0.03 -0.03 0.17 0.25 0.16 0.56 0.63 0.37 1.8
Q7_13 -0.08 -0.13 0.02 -0.04 0.26 0.09 0.72 0.59 0.41 1.4
Q7_14 -0.03 0.18 -0.05 -0.11 0.57 -0.14 0.35 0.52 0.48 2.2
Q7_15 0.12 0.01 0.03 0.82 -0.01 -0.16 0.00 0.64 0.36 1.1
PA3 PA4 PA2 PA5 PA6 PA1 PA7
SS loadings 8.79 5.81 5.50 5.13 5.28 4.72 1.75
Proportion Var 0.15 0.10 0.10 0.09 0.09 0.08 0.03
Cumulative Var 0.15 0.26 0.35 0.44 0.54 0.62 0.65
Proportion Explained 0.24 0.16 0.15 0.14 0.14 0.13 0.05
Cumulative Proportion 0.24 0.39 0.54 0.68 0.83 0.95 1.00
With factor correlations of
PA3 PA4 PA2 PA5 PA6 PA1 PA7
PA3 1.00 0.62 0.52 0.66 0.55 0.61 0.20
PA4 0.62 1.00 0.52 0.66 0.61 0.59 0.30
PA2 0.52 0.52 1.00 0.63 0.46 0.65 0.04
PA5 0.66 0.66 0.63 1.00 0.63 0.75 0.22
PA6 0.55 0.61 0.46 0.63 1.00 0.55 0.28
PA1 0.61 0.59 0.65 0.75 0.55 1.00 0.13
PA7 0.20 0.30 0.04 0.22 0.28 0.13 1.00
Mean item complexity = 1.9
Test of the hypothesis that 7 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1218 and the objective function was 37.91
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.04
Fit based upon off diagonal values = 1print(fit$loadings, cutoff=0.3)
Loadings:
PA3 PA4 PA2 PA5 PA6 PA1 PA7
Q4_1 0.876
Q4_2 0.769
Q4_3 0.881
Q4_4 0.787
Q4_5 0.474
Q4_6 0.326
Q4_7 0.411
Q4_8 0.786
Q4_9 0.585
Q4_10 0.711
Q4_11 0.569
Q4_12 -0.315 0.697
Q4_13 0.342 0.768
Q4_14 0.354 0.472
Q4_15 0.555 0.381
Q4_16 0.552
Q4_17 0.585
Q4_18 0.778
Q4_19 0.547 0.384
Q5_1 0.396 0.311
Q5_2 0.665
Q5_3 0.750 0.325
Q5_4 0.931
Q5_5 0.890 -0.331
Q5_6 0.798
Q5_7 0.336 0.300
Q5_8 0.766
Q5_9 0.530
Q5_10 0.375
Q5_11 0.325
Q5_12 0.427
Q6_1 0.815 -0.327
Q6_2 0.991
Q6_3 0.952
Q6_4 0.907
Q6_5
Q6_6 0.572
Q6_7 0.606
Q6_8 0.645
Q6_9 0.646
Q6_10
Q6_11
Q7_1 0.639
Q7_2 0.425
Q7_3 0.848
Q7_4 0.503 0.628
Q7_5 0.348 0.553
Q7_6 0.514
Q7_7 0.766
Q7_8 0.568
Q7_9 0.837
Q7_10 0.868
Q7_11 0.574 0.347
Q7_12 0.556
Q7_13 0.718
Q7_14 0.568 0.353
Q7_15 0.815
PA3 PA4 PA2 PA5 PA6 PA1 PA7
SS loadings 7.292 5.403 5.036 4.454 4.354 3.879 1.550
Proportion Var 0.128 0.095 0.088 0.078 0.076 0.068 0.027
Cumulative Var 0.128 0.223 0.311 0.389 0.466 0.534 0.561fit <- psych::fa(r = cov.dat, nfactors = 7, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 7, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA4 PA5 PA3 PA6 PA2 PA7 h2 u2 H2 U2
Q4_1 0.70 0.02 0.24 -0.01 -0.24 -0.12 0.06 0.48 0.27 0.64 0.36
Q4_2 0.66 0.01 0.12 -0.06 -0.07 -0.05 0.00 0.43 0.24 0.64 0.36
Q4_3 0.68 0.01 0.17 -0.01 -0.22 -0.05 0.09 0.45 0.25 0.65 0.35
Q4_4 0.58 -0.01 0.17 0.00 -0.26 0.18 -0.06 0.44 0.22 0.67 0.33
Q4_5 0.46 0.08 0.16 0.01 0.02 -0.07 -0.05 0.41 0.37 0.52 0.48
Q4_6 0.27 0.27 0.11 0.18 -0.15 0.10 0.06 0.44 0.53 0.45 0.55
Q4_7 0.14 0.26 -0.10 0.12 -0.11 0.36 -0.01 0.32 0.56 0.37 0.63
Q4_8 0.77 -0.07 -0.04 0.03 0.04 -0.04 -0.04 0.53 0.33 0.62 0.38
Q4_9 0.68 -0.12 -0.09 -0.05 0.17 0.12 0.00 0.50 0.52 0.49 0.51
Q4_10 0.54 -0.02 0.00 0.05 -0.13 0.19 -0.02 0.35 0.27 0.56 0.44
Q4_11 0.61 -0.04 0.00 -0.01 0.19 0.06 0.03 0.56 0.36 0.61 0.39
Q4_12 0.47 -0.10 -0.31 0.18 0.54 0.10 -0.07 0.65 0.46 0.59 0.41
Q4_13 0.44 -0.06 -0.15 0.12 0.41 -0.15 0.00 0.40 0.37 0.52 0.48
Q4_14 0.40 0.05 -0.09 0.04 0.37 -0.03 0.13 0.44 0.36 0.55 0.45
Q4_15 0.59 0.08 -0.03 -0.03 0.23 -0.08 0.00 0.53 0.31 0.63 0.37
Q4_16 0.55 0.23 -0.03 0.12 -0.02 -0.13 -0.01 0.47 0.41 0.53 0.47
Q4_17 0.63 -0.13 -0.03 0.05 0.13 0.06 -0.07 0.47 0.46 0.51 0.49
Q4_18 0.69 0.03 0.03 -0.08 0.03 -0.02 -0.08 0.50 0.18 0.74 0.26
Q4_19 0.64 0.00 -0.09 -0.07 0.24 0.12 -0.04 0.60 0.34 0.64 0.36
Q5_1 0.17 0.33 0.22 -0.20 0.27 -0.11 0.05 0.46 0.45 0.51 0.49
Q5_2 0.01 0.66 -0.05 0.01 0.10 0.07 -0.08 0.57 0.50 0.53 0.47
Q5_3 0.05 0.72 0.06 -0.11 0.29 -0.24 -0.02 0.68 0.41 0.62 0.38
Q5_4 0.00 1.04 -0.14 0.05 -0.34 0.16 0.02 0.81 0.41 0.66 0.34
Q5_5 0.00 0.96 -0.09 0.01 -0.41 0.14 0.03 0.66 0.41 0.62 0.38
Q5_6 0.04 0.70 0.03 -0.03 -0.04 0.04 -0.07 0.54 0.28 0.66 0.34
Q5_7 -0.04 0.30 -0.06 0.16 0.27 0.12 -0.07 0.42 0.46 0.48 0.52
Q5_8 -0.10 0.75 0.08 -0.01 0.04 0.07 -0.08 0.68 0.43 0.61 0.39
Q5_9 0.04 0.46 0.28 0.03 0.16 -0.13 -0.05 0.62 0.37 0.62 0.38
Q5_10 0.07 0.29 0.08 -0.02 0.11 0.08 -0.14 0.30 0.40 0.43 0.57
Q5_11 0.13 0.19 0.35 0.02 0.13 -0.09 0.21 0.46 0.45 0.51 0.49
Q5_12 0.00 0.36 0.19 0.05 0.03 0.13 0.00 0.43 0.48 0.47 0.53
Q6_1 0.01 0.01 0.05 0.65 0.08 -0.21 0.14 0.44 0.27 0.62 0.38
Q6_2 -0.04 0.06 0.02 0.88 -0.12 -0.05 0.07 0.63 0.20 0.76 0.24
Q6_3 -0.04 0.09 -0.09 0.88 -0.10 0.00 0.03 0.61 0.24 0.72 0.28
Q6_4 -0.01 0.03 -0.04 0.81 -0.14 0.03 -0.06 0.53 0.33 0.62 0.38
Q6_5 -0.02 -0.04 0.28 0.19 0.28 -0.09 0.20 0.32 0.74 0.30 0.70
Q6_6 0.01 -0.10 0.17 0.48 0.09 0.03 -0.09 0.43 0.24 0.64 0.36
Q6_7 0.09 -0.14 0.06 0.50 0.20 0.06 -0.05 0.53 0.24 0.69 0.31
Q6_8 -0.02 -0.18 0.16 0.54 0.20 0.05 -0.09 0.56 0.26 0.68 0.32
Q6_9 0.04 -0.25 -0.04 -0.06 0.97 0.16 0.33 0.72 0.77 0.48 0.52
Q6_10 -0.02 0.01 0.22 0.03 0.36 0.06 0.14 0.34 0.50 0.40 0.60
Q6_11 -0.03 0.11 0.07 0.15 0.33 0.26 0.03 0.53 0.41 0.56 0.44
Q7_1 -0.02 0.11 0.58 0.08 0.03 0.00 0.04 0.54 0.28 0.66 0.34
Q7_2 0.00 0.09 0.25 0.03 0.04 0.32 -0.05 0.40 0.34 0.54 0.46
Q7_3 -0.05 0.13 -0.19 0.05 0.07 0.81 0.10 0.67 0.35 0.66 0.34
Q7_4 -0.09 -0.08 0.51 0.05 -0.19 0.57 -0.18 0.60 0.36 0.63 0.37
Q7_5 0.06 -0.13 0.36 -0.07 0.14 0.49 0.00 0.60 0.30 0.66 0.34
Q7_6 0.08 -0.04 0.01 0.05 0.25 0.47 0.06 0.47 0.37 0.56 0.44
Q7_7 0.09 0.14 -0.21 -0.11 0.16 0.74 0.18 0.70 0.33 0.68 0.32
Q7_8 0.05 -0.08 0.23 -0.14 0.17 0.45 -0.04 0.42 0.35 0.55 0.45
Q7_9 0.08 -0.02 0.85 -0.02 -0.08 -0.05 0.03 0.64 0.35 0.65 0.35
Q7_10 0.15 -0.10 0.82 0.07 -0.14 -0.06 -0.06 0.61 0.29 0.68 0.32
Q7_11 0.11 -0.06 0.48 -0.04 -0.09 0.25 0.05 0.39 0.31 0.56 0.44
Q7_12 -0.01 0.02 0.19 0.00 0.28 0.23 0.49 0.56 0.49 0.53 0.47
Q7_13 -0.07 -0.08 -0.07 0.02 0.41 0.19 0.69 0.54 0.44 0.55 0.45
Q7_14 -0.05 0.22 -0.12 -0.01 0.04 0.52 0.29 0.46 0.51 0.47 0.53
Q7_15 0.07 -0.04 0.74 0.00 -0.08 -0.04 -0.01 0.47 0.35 0.57 0.43
PA1 PA4 PA5 PA3 PA6 PA2 PA7
SS loadings 7.37 5.16 4.31 3.89 3.38 4.08 1.10
Proportion Var 0.15 0.10 0.08 0.08 0.07 0.08 0.02
Cumulative Var 0.15 0.25 0.33 0.41 0.47 0.56 0.58
Proportion Explained 0.25 0.18 0.15 0.13 0.12 0.14 0.04
Cumulative Proportion 0.25 0.43 0.57 0.71 0.82 0.96 1.00
Standardized loadings (pattern matrix)
item PA1 PA4 PA5 PA3 PA6 PA2 PA7 h2 u2
Q4_1 1 0.80 0.02 0.27 -0.01 -0.27 -0.13 0.07 0.64 0.36
Q4_2 2 0.80 0.01 0.15 -0.08 -0.08 -0.06 -0.01 0.64 0.36
Q4_3 3 0.82 0.01 0.21 -0.01 -0.26 -0.06 0.10 0.65 0.35
Q4_4 4 0.71 -0.01 0.21 0.00 -0.31 0.22 -0.07 0.67 0.33
Q4_5 5 0.53 0.09 0.18 0.02 0.02 -0.08 -0.06 0.52 0.48
Q4_6 6 0.27 0.27 0.11 0.18 -0.15 0.11 0.07 0.45 0.55
Q4_7 7 0.15 0.27 -0.11 0.13 -0.12 0.38 -0.01 0.37 0.63
Q4_8 8 0.83 -0.08 -0.04 0.03 0.05 -0.04 -0.05 0.62 0.38
Q4_9 9 0.68 -0.11 -0.09 -0.05 0.17 0.12 0.00 0.49 0.51
Q4_10 10 0.68 -0.03 0.00 0.07 -0.17 0.24 -0.02 0.56 0.44
Q4_11 11 0.64 -0.04 0.00 -0.01 0.20 0.07 0.04 0.61 0.39
Q4_12 12 0.44 -0.09 -0.30 0.17 0.51 0.10 -0.06 0.59 0.41
Q4_13 13 0.50 -0.07 -0.17 0.14 0.46 -0.17 0.00 0.52 0.48
Q4_14 14 0.44 0.06 -0.10 0.04 0.42 -0.03 0.15 0.55 0.45
Q4_15 15 0.65 0.08 -0.03 -0.03 0.25 -0.08 0.00 0.63 0.37
Q4_16 16 0.58 0.24 -0.03 0.13 -0.02 -0.14 -0.01 0.53 0.47
Q4_17 17 0.66 -0.13 -0.03 0.05 0.13 0.06 -0.07 0.51 0.49
Q4_18 18 0.84 0.03 0.04 -0.10 0.04 -0.02 -0.10 0.74 0.26
Q4_19 19 0.66 0.00 -0.09 -0.07 0.24 0.13 -0.05 0.64 0.36
Q5_1 20 0.18 0.34 0.23 -0.21 0.28 -0.12 0.05 0.51 0.49
Q5_2 21 0.01 0.64 -0.05 0.01 0.10 0.07 -0.07 0.53 0.47
Q5_3 22 0.05 0.69 0.06 -0.10 0.27 -0.23 -0.02 0.62 0.38
Q5_4 23 0.00 0.94 -0.13 0.05 -0.31 0.15 0.02 0.66 0.34
Q5_5 24 0.00 0.93 -0.08 0.01 -0.39 0.13 0.03 0.62 0.38
Q5_6 25 0.04 0.78 0.03 -0.03 -0.05 0.04 -0.07 0.66 0.34
Q5_7 26 -0.04 0.32 -0.07 0.17 0.29 0.13 -0.07 0.48 0.52
Q5_8 27 -0.09 0.71 0.08 -0.01 0.04 0.07 -0.07 0.61 0.39
Q5_9 28 0.04 0.46 0.28 0.03 0.16 -0.13 -0.05 0.62 0.38
Q5_10 29 0.08 0.34 0.10 -0.03 0.13 0.10 -0.17 0.43 0.57
Q5_11 30 0.14 0.19 0.37 0.03 0.14 -0.09 0.22 0.51 0.49
Q5_12 31 0.00 0.38 0.20 0.05 0.03 0.14 0.00 0.47 0.53
Q6_1 32 0.01 0.02 0.06 0.77 0.10 -0.25 0.16 0.62 0.38
Q6_2 33 -0.04 0.06 0.02 0.97 -0.13 -0.06 0.07 0.76 0.24
Q6_3 34 -0.04 0.09 -0.10 0.95 -0.11 0.00 0.03 0.72 0.28
Q6_4 35 -0.02 0.03 -0.05 0.88 -0.16 0.03 -0.07 0.62 0.38
Q6_5 36 -0.01 -0.04 0.27 0.18 0.27 -0.09 0.20 0.30 0.70
Q6_6 37 0.01 -0.12 0.21 0.58 0.11 0.03 -0.11 0.64 0.36
Q6_7 38 0.11 -0.16 0.07 0.57 0.23 0.07 -0.06 0.69 0.31
Q6_8 39 -0.03 -0.20 0.18 0.60 0.23 0.06 -0.09 0.68 0.32
Q6_9 40 0.03 -0.21 -0.04 -0.05 0.80 0.13 0.27 0.48 0.52
Q6_10 41 -0.03 0.01 0.25 0.03 0.39 0.06 0.15 0.40 0.60
Q6_11 42 -0.03 0.12 0.07 0.15 0.34 0.26 0.03 0.56 0.44
Q7_1 43 -0.03 0.13 0.65 0.09 0.04 0.00 0.05 0.66 0.34
Q7_2 44 0.00 0.10 0.29 0.03 0.05 0.38 -0.06 0.54 0.46
Q7_3 45 -0.05 0.13 -0.19 0.05 0.07 0.80 0.10 0.66 0.34
Q7_4 46 -0.09 -0.08 0.52 0.05 -0.20 0.58 -0.18 0.63 0.37
Q7_5 47 0.06 -0.14 0.38 -0.08 0.14 0.51 0.00 0.66 0.34
Q7_6 48 0.09 -0.04 0.01 0.05 0.27 0.51 0.07 0.56 0.44
Q7_7 49 0.09 0.14 -0.21 -0.10 0.16 0.73 0.18 0.68 0.32
Q7_8 50 0.06 -0.09 0.26 -0.16 0.20 0.51 -0.04 0.55 0.45
Q7_9 51 0.08 -0.02 0.86 -0.02 -0.08 -0.05 0.03 0.65 0.35
Q7_10 52 0.16 -0.11 0.87 0.08 -0.15 -0.07 -0.06 0.68 0.32
Q7_11 53 0.14 -0.07 0.58 -0.05 -0.11 0.29 0.06 0.56 0.44
Q7_12 54 -0.01 0.02 0.18 0.00 0.28 0.23 0.48 0.53 0.47
Q7_13 55 -0.07 -0.08 -0.07 0.02 0.42 0.19 0.70 0.55 0.45
Q7_14 56 -0.05 0.22 -0.12 -0.01 0.04 0.52 0.30 0.47 0.53
Q7_15 57 0.08 -0.04 0.82 0.00 -0.09 -0.05 -0.01 0.57 0.43
PA1 PA4 PA5 PA3 PA6 PA2 PA7
SS loadings 8.27 5.80 4.84 4.36 3.80 4.58 1.24
Proportion Var 0.15 0.10 0.08 0.08 0.07 0.08 0.02
Cumulative Var 0.15 0.25 0.33 0.41 0.47 0.56 0.58
Cum. factor Var 0.25 0.43 0.57 0.71 0.82 0.96 1.00
With factor correlations of
PA1 PA4 PA5 PA3 PA6 PA2 PA7
PA1 1.00 0.63 0.71 0.54 0.62 0.52 -0.05
PA4 0.63 1.00 0.71 0.51 0.63 0.58 0.00
PA5 0.71 0.71 1.00 0.63 0.73 0.62 -0.05
PA3 0.54 0.51 0.63 1.00 0.66 0.38 -0.22
PA6 0.62 0.63 0.73 0.66 1.00 0.48 -0.25
PA2 0.52 0.58 0.62 0.38 0.48 1.00 0.12
PA7 -0.05 0.00 -0.05 -0.22 -0.25 0.12 1.00
Mean item complexity = 1.8
Test of the hypothesis that 7 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1218 and the objective function was 8.01
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.03
Fit based upon off diagonal values = 0.99
Measures of factor score adequacy
PA1 PA4 PA5 PA3 PA6 PA2
Correlation of (regression) scores with factors 0.98 0.97 0.97 0.97 0.94 0.95
Multiple R square of scores with factors 0.96 0.93 0.93 0.94 0.89 0.91
Minimum correlation of possible factor scores 0.91 0.87 0.87 0.88 0.78 0.82
PA7
Correlation of (regression) scores with factors 0.85
Multiple R square of scores with factors 0.72
Minimum correlation of possible factor scores 0.45print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA4 PA5 PA3 PA6 PA2 PA7
Q4_1 0.696
Q4_2 0.659
Q4_3 0.684
Q4_4 0.581
Q4_5 0.463
Q4_6
Q4_7 0.358
Q4_8 0.767
Q4_9 0.683
Q4_10 0.539
Q4_11 0.611
Q4_12 0.466 -0.311 0.536
Q4_13 0.441 0.407
Q4_14 0.397 0.374
Q4_15 0.593
Q4_16 0.545
Q4_17 0.633
Q4_18 0.688
Q4_19 0.642
Q5_1 0.327
Q5_2 0.657
Q5_3 0.720
Q5_4 1.039 -0.340
Q5_5 0.964 -0.407
Q5_6 0.703
Q5_7 0.302
Q5_8 0.754
Q5_9 0.456
Q5_10
Q5_11 0.352
Q5_12 0.362
Q6_1 0.650
Q6_2 0.884
Q6_3 0.876
Q6_4 0.811
Q6_5
Q6_6 0.478
Q6_7 0.502
Q6_8 0.541
Q6_9 0.974 0.331
Q6_10 0.357
Q6_11 0.327
Q7_1 0.583
Q7_2 0.325
Q7_3 0.809
Q7_4 0.510 0.568
Q7_5 0.358 0.487
Q7_6 0.466
Q7_7 0.744
Q7_8 0.449
Q7_9 0.850
Q7_10 0.818
Q7_11 0.483
Q7_12 0.492
Q7_13 0.414 0.694
Q7_14 0.515
Q7_15 0.738
PA1 PA4 PA5 PA3 PA6 PA2 PA7
SS loadings 6.428 5.245 4.021 3.747 3.475 3.388 1.268
Proportion Var 0.113 0.092 0.071 0.066 0.061 0.059 0.022
Cumulative Var 0.113 0.205 0.275 0.341 0.402 0.461 0.484Eight factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 8, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 8, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA3 PA4 PA2 PA6 PA1 PA5 PA7 PA8 h2 u2 com
Q4_1 0.86 -0.03 -0.01 -0.17 -0.17 0.23 0.07 0.15 0.74 0.26 1.4
Q4_2 0.77 -0.02 -0.14 -0.07 0.10 0.18 -0.04 0.13 0.73 0.27 1.3
Q4_3 0.89 0.03 0.03 -0.08 -0.25 0.13 0.12 0.02 0.75 0.25 1.3
Q4_4 0.78 -0.04 0.00 0.24 -0.24 0.17 -0.08 0.14 0.78 0.22 1.6
Q4_5 0.47 0.04 -0.07 -0.07 0.24 0.23 -0.11 0.17 0.62 0.38 2.6
Q4_6 0.30 0.09 0.09 0.01 -0.04 0.20 0.05 0.51 0.69 0.31 2.2
Q4_7 0.12 0.13 0.08 0.32 -0.03 -0.03 -0.02 0.41 0.50 0.50 2.4
Q4_8 0.79 -0.08 0.03 -0.02 0.19 -0.06 -0.06 0.03 0.69 0.31 1.2
Q4_9 0.63 0.01 0.03 0.16 0.16 -0.15 0.06 -0.19 0.58 0.42 1.6
Q4_10 0.71 -0.04 0.09 0.26 -0.09 -0.06 -0.03 0.11 0.67 0.33 1.4
Q4_11 0.58 -0.04 0.00 0.09 0.28 -0.02 0.05 0.03 0.67 0.33 1.5
Q4_12 0.30 -0.13 0.13 0.10 0.74 -0.27 -0.05 0.04 0.70 0.30 1.8
Q4_13 0.37 -0.05 0.10 -0.19 0.73 -0.11 0.00 -0.12 0.72 0.28 1.8
Q4_14 0.36 0.02 0.09 -0.10 0.50 -0.08 0.18 0.05 0.63 0.37 2.4
Q4_15 0.58 0.15 0.02 -0.04 0.31 -0.07 0.01 -0.09 0.70 0.30 1.8
Q4_16 0.56 0.25 0.19 -0.18 0.07 -0.06 0.00 0.05 0.60 0.40 2.0
Q4_17 0.60 -0.09 0.04 0.10 0.24 -0.05 -0.08 -0.01 0.57 0.43 1.5
Q4_18 0.78 0.05 -0.14 0.01 0.23 0.03 -0.13 0.05 0.84 0.16 1.3
Q4_19 0.57 0.06 -0.02 0.15 0.31 -0.12 0.01 -0.08 0.70 0.30 1.9
Q5_1 0.15 0.40 -0.19 -0.07 0.28 0.22 0.05 -0.08 0.57 0.43 3.6
Q5_2 -0.04 0.59 -0.01 -0.01 0.23 0.00 -0.05 0.14 0.57 0.43 1.4
Q5_3 -0.02 0.71 -0.09 -0.24 0.33 0.10 -0.02 0.00 0.69 0.31 1.8
Q5_4 0.01 0.96 0.00 0.10 -0.27 -0.13 0.02 0.09 0.70 0.30 1.2
Q5_5 0.05 1.02 0.03 0.10 -0.46 -0.13 0.04 -0.03 0.68 0.32 1.5
Q5_6 0.06 0.84 0.01 0.04 -0.07 0.00 -0.05 -0.02 0.71 0.29 1.0
Q5_7 -0.08 0.35 0.24 0.20 0.28 -0.11 -0.07 -0.03 0.53 0.47 4.0
Q5_8 -0.11 0.77 0.01 0.11 0.04 0.04 -0.06 0.01 0.66 0.34 1.1
Q5_9 0.03 0.48 0.04 -0.07 0.19 0.26 -0.08 0.06 0.68 0.32 2.1
Q5_10 0.06 0.38 -0.01 0.17 0.21 0.07 -0.18 -0.05 0.50 0.50 2.7
Q5_11 0.13 0.26 0.05 -0.05 0.06 0.30 0.24 -0.02 0.56 0.44 3.6
Q5_12 -0.03 0.42 0.06 0.14 0.03 0.18 0.01 0.04 0.51 0.49 1.7
Q6_1 0.00 -0.01 0.81 -0.33 0.13 0.12 0.19 0.01 0.77 0.23 1.6
Q6_2 -0.04 0.07 0.98 -0.12 -0.09 0.00 0.08 0.11 0.83 0.17 1.1
Q6_3 -0.01 0.10 0.95 -0.01 -0.04 -0.13 -0.01 0.07 0.81 0.19 1.1
Q6_4 -0.01 0.07 0.94 0.01 -0.12 -0.11 -0.05 0.00 0.71 0.29 1.1
Q6_5 -0.03 -0.02 0.23 -0.03 0.20 0.26 0.16 -0.06 0.36 0.64 3.8
Q6_6 -0.02 -0.11 0.56 0.16 0.18 0.23 -0.17 -0.01 0.77 0.23 2.1
Q6_7 0.08 -0.14 0.61 0.15 0.28 0.02 -0.07 -0.03 0.79 0.21 1.8
Q6_8 -0.07 -0.15 0.65 0.16 0.23 0.13 -0.09 -0.05 0.77 0.23 1.7
Q6_9 -0.07 -0.25 -0.03 0.18 0.72 0.02 0.28 0.00 0.54 0.46 1.7
Q6_10 -0.06 -0.06 0.02 0.09 0.35 0.29 0.16 0.13 0.45 0.55 3.0
Q6_11 -0.08 -0.03 0.10 0.25 0.43 0.15 0.03 0.28 0.68 0.32 3.0
Q7_1 -0.02 0.13 0.10 0.09 0.02 0.61 0.06 0.04 0.74 0.26 1.2
Q7_2 0.01 0.12 0.06 0.46 0.04 0.22 -0.01 -0.01 0.59 0.41 1.7
Q7_3 -0.08 0.10 0.02 0.75 0.06 -0.22 0.18 0.18 0.69 0.31 1.5
Q7_4 -0.07 -0.05 0.05 0.68 -0.20 0.43 -0.11 0.09 0.66 0.34 2.1
Q7_5 0.06 -0.09 -0.05 0.58 0.10 0.28 0.08 0.00 0.70 0.30 1.7
Q7_6 0.07 0.03 0.10 0.50 0.19 -0.06 0.17 -0.05 0.61 0.39 1.8
Q7_7 0.06 0.14 -0.18 0.66 0.17 -0.23 0.30 0.12 0.77 0.23 2.2
Q7_8 0.03 0.03 -0.13 0.64 0.09 0.18 0.03 -0.17 0.63 0.37 1.5
Q7_9 0.11 0.02 -0.01 0.06 -0.10 0.78 0.05 0.00 0.71 0.29 1.1
Q7_10 0.19 -0.06 0.05 0.10 -0.15 0.81 -0.08 0.03 0.77 0.23 1.3
Q7_11 0.20 0.07 0.05 0.48 -0.32 0.44 0.11 -0.26 0.75 0.25 4.0
Q7_12 -0.01 0.06 0.04 0.20 0.09 0.10 0.61 -0.13 0.67 0.33 1.4
Q7_13 -0.08 -0.14 0.00 0.13 0.14 -0.03 0.73 0.08 0.58 0.42 1.3
Q7_14 -0.04 0.15 -0.07 0.45 -0.06 -0.09 0.37 0.19 0.53 0.47 2.8
Q7_15 0.10 -0.09 -0.04 0.07 -0.07 0.83 -0.03 0.14 0.67 0.33 1.1
PA3 PA4 PA2 PA6 PA1 PA5 PA7 PA8
SS loadings 8.93 5.64 5.35 5.24 4.80 4.67 1.95 1.22
Proportion Var 0.16 0.10 0.09 0.09 0.08 0.08 0.03 0.02
Cumulative Var 0.16 0.26 0.35 0.44 0.53 0.61 0.64 0.66
Proportion Explained 0.24 0.15 0.14 0.14 0.13 0.12 0.05 0.03
Cumulative Proportion 0.24 0.39 0.53 0.67 0.79 0.92 0.97 1.00
With factor correlations of
PA3 PA4 PA2 PA6 PA1 PA5 PA7 PA8
PA3 1.00 0.61 0.52 0.55 0.63 0.66 0.21 0.11
PA4 0.61 1.00 0.50 0.60 0.66 0.67 0.30 0.28
PA2 0.52 0.50 1.00 0.45 0.67 0.63 0.04 0.12
PA6 0.55 0.60 0.45 1.00 0.57 0.58 0.34 0.21
PA1 0.63 0.66 0.67 0.57 1.00 0.74 0.17 0.14
PA5 0.66 0.67 0.63 0.58 0.74 1.00 0.25 0.12
PA7 0.21 0.30 0.04 0.34 0.17 0.25 1.00 0.13
PA8 0.11 0.28 0.12 0.21 0.14 0.12 0.13 1.00
Mean item complexity = 1.9
Test of the hypothesis that 8 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1168 and the objective function was 37.08
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.04
Fit based upon off diagonal values = 1print(fit$loadings, cutoff=0.3)
Loadings:
PA3 PA4 PA2 PA6 PA1 PA5 PA7 PA8
Q4_1 0.865
Q4_2 0.766
Q4_3 0.888
Q4_4 0.779
Q4_5 0.465
Q4_6 0.509
Q4_7 0.320 0.411
Q4_8 0.794
Q4_9 0.628
Q4_10 0.710
Q4_11 0.579
Q4_12 0.304 0.743
Q4_13 0.368 0.726
Q4_14 0.360 0.502
Q4_15 0.576 0.305
Q4_16 0.555
Q4_17 0.597
Q4_18 0.783
Q4_19 0.572 0.315
Q5_1 0.395
Q5_2 0.588
Q5_3 0.706 0.327
Q5_4 0.961
Q5_5 1.023 -0.455
Q5_6 0.840
Q5_7 0.346
Q5_8 0.771
Q5_9 0.478
Q5_10 0.381
Q5_11
Q5_12 0.419
Q6_1 0.810 -0.330
Q6_2 0.976
Q6_3 0.951
Q6_4 0.941
Q6_5
Q6_6 0.561
Q6_7 0.606
Q6_8 0.647
Q6_9 0.718
Q6_10 0.353
Q6_11 0.431
Q7_1 0.606
Q7_2 0.459
Q7_3 0.746
Q7_4 0.681 0.435
Q7_5 0.584
Q7_6 0.500
Q7_7 0.659
Q7_8 0.642
Q7_9 0.777
Q7_10 0.809
Q7_11 0.478 -0.318 0.440
Q7_12 0.615
Q7_13 0.731
Q7_14 0.451 0.373
Q7_15 0.827
PA3 PA4 PA2 PA6 PA1 PA5 PA7 PA8
SS loadings 7.439 5.444 4.910 4.094 4.060 3.933 1.689 0.998
Proportion Var 0.131 0.096 0.086 0.072 0.071 0.069 0.030 0.018
Cumulative Var 0.131 0.226 0.312 0.384 0.455 0.524 0.554 0.571fit <- psych::fa(r = cov.dat, nfactors = 8, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 8, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8 h2 u2 H2 U2
Q4_1 0.66 -0.02 0.25 -0.22 -0.03 -0.12 0.06 0.16 0.48 0.27 0.65 0.35
Q4_2 0.63 -0.03 0.13 -0.05 -0.08 -0.05 -0.01 0.11 0.43 0.24 0.65 0.35
Q4_3 0.67 0.05 0.17 -0.24 -0.01 -0.06 0.09 0.05 0.45 0.25 0.65 0.35
Q4_4 0.56 -0.02 0.17 -0.25 -0.01 0.17 -0.06 0.12 0.44 0.22 0.67 0.33
Q4_5 0.43 -0.01 0.18 0.06 -0.01 -0.06 -0.06 0.15 0.42 0.36 0.54 0.46
Q4_6 0.20 0.02 0.18 -0.03 0.10 0.11 0.05 0.50 0.60 0.36 0.63 0.37
Q4_7 0.09 0.08 -0.07 -0.02 0.06 0.38 -0.03 0.36 0.40 0.48 0.46 0.54
Q4_8 0.76 -0.06 -0.04 0.03 0.03 -0.04 -0.04 0.02 0.53 0.33 0.62 0.38
Q4_9 0.73 0.02 -0.14 0.08 0.00 0.11 0.01 -0.18 0.54 0.47 0.54 0.46
Q4_10 0.52 -0.03 0.00 -0.13 0.04 0.19 -0.02 0.10 0.35 0.27 0.56 0.44
Q4_11 0.60 -0.04 0.00 0.19 -0.01 0.06 0.03 0.04 0.56 0.36 0.61 0.39
Q4_12 0.47 -0.13 -0.30 0.55 0.18 0.10 -0.07 0.01 0.65 0.46 0.58 0.42
Q4_13 0.46 -0.03 -0.15 0.38 0.14 -0.15 0.01 -0.09 0.41 0.36 0.53 0.47
Q4_14 0.39 0.01 -0.08 0.40 0.03 -0.03 0.12 0.05 0.44 0.36 0.55 0.45
Q4_15 0.60 0.11 -0.04 0.20 -0.01 -0.08 0.00 -0.06 0.53 0.31 0.64 0.36
Q4_16 0.53 0.21 -0.02 -0.02 0.12 -0.13 -0.02 0.08 0.47 0.42 0.53 0.47
Q4_17 0.63 -0.10 -0.03 0.10 0.06 0.06 -0.07 -0.01 0.47 0.46 0.51 0.49
Q4_18 0.67 0.00 0.04 0.04 -0.09 -0.01 -0.08 0.06 0.50 0.18 0.74 0.26
Q4_19 0.66 0.05 -0.10 0.20 -0.04 0.12 -0.04 -0.07 0.61 0.33 0.65 0.35
Q5_1 0.18 0.32 0.22 0.27 -0.19 -0.10 0.05 -0.04 0.46 0.45 0.51 0.49
Q5_2 -0.01 0.55 -0.03 0.16 -0.02 0.08 -0.09 0.15 0.58 0.49 0.54 0.46
Q5_3 0.05 0.65 0.07 0.31 -0.11 -0.23 -0.03 0.02 0.68 0.41 0.62 0.38
Q5_4 0.01 1.09 -0.16 -0.39 0.07 0.14 0.02 0.04 0.82 0.40 0.67 0.33
Q5_5 0.03 1.15 -0.13 -0.54 0.07 0.10 0.05 -0.10 0.75 0.32 0.70 0.30
Q5_6 0.05 0.72 0.02 -0.07 -0.02 0.03 -0.06 -0.02 0.54 0.28 0.66 0.34
Q5_7 -0.02 0.31 -0.07 0.26 0.18 0.12 -0.06 -0.05 0.42 0.46 0.48 0.52
Q5_8 -0.09 0.74 0.08 0.04 0.00 0.07 -0.07 0.01 0.68 0.44 0.61 0.39
Q5_9 0.03 0.37 0.30 0.20 0.01 -0.12 -0.06 0.08 0.62 0.37 0.63 0.37
Q5_10 0.07 0.28 0.08 0.11 -0.02 0.09 -0.14 -0.02 0.30 0.40 0.43 0.57
Q5_11 0.13 0.19 0.34 0.13 0.03 -0.09 0.21 0.00 0.46 0.45 0.51 0.49
Q5_12 -0.01 0.34 0.19 0.04 0.04 0.13 0.00 0.05 0.43 0.48 0.47 0.53
Q6_1 0.00 0.02 0.05 0.08 0.64 -0.21 0.14 0.04 0.44 0.28 0.61 0.39
Q6_2 -0.05 0.05 0.02 -0.12 0.86 -0.06 0.06 0.11 0.63 0.20 0.76 0.24
Q6_3 -0.04 0.11 -0.09 -0.10 0.86 -0.01 0.03 0.07 0.61 0.24 0.72 0.28
Q6_4 0.00 0.08 -0.05 -0.18 0.81 0.02 -0.06 0.01 0.54 0.32 0.63 0.37
Q6_5 0.00 0.00 0.26 0.26 0.21 -0.09 0.21 -0.07 0.33 0.74 0.31 0.69
Q6_6 0.02 -0.07 0.16 0.07 0.49 0.03 -0.08 -0.04 0.43 0.24 0.65 0.35
Q6_7 0.11 -0.11 0.05 0.18 0.51 0.06 -0.05 -0.05 0.53 0.24 0.69 0.31
Q6_8 -0.01 -0.15 0.15 0.18 0.55 0.06 -0.08 -0.07 0.56 0.25 0.69 0.31
Q6_9 0.05 -0.30 -0.04 1.00 -0.06 0.17 0.33 -0.03 0.72 0.78 0.48 0.52
Q6_10 -0.04 -0.07 0.24 0.40 0.01 0.07 0.14 0.07 0.34 0.49 0.41 0.59
Q6_11 -0.07 -0.06 0.10 0.44 0.09 0.27 0.01 0.23 0.58 0.36 0.62 0.38
Q7_1 -0.03 0.09 0.58 0.05 0.08 0.01 0.04 0.03 0.54 0.28 0.66 0.34
Q7_2 0.01 0.12 0.23 0.02 0.05 0.32 -0.04 -0.04 0.40 0.33 0.55 0.45
Q7_3 -0.06 0.09 -0.19 0.10 0.03 0.79 0.10 0.14 0.67 0.35 0.66 0.34
Q7_4 -0.10 -0.11 0.50 -0.17 0.05 0.58 -0.17 0.08 0.60 0.36 0.63 0.37
Q7_5 0.07 -0.11 0.34 0.13 -0.06 0.49 0.01 -0.02 0.60 0.30 0.66 0.34
Q7_6 0.10 0.00 -0.01 0.22 0.06 0.46 0.07 -0.05 0.48 0.37 0.57 0.43
Q7_7 0.09 0.15 -0.22 0.16 -0.10 0.72 0.18 0.05 0.70 0.33 0.68 0.32
Q7_8 0.08 0.01 0.20 0.12 -0.10 0.45 -0.02 -0.15 0.44 0.33 0.58 0.42
Q7_9 0.08 -0.03 0.84 -0.06 -0.02 -0.03 0.03 0.02 0.64 0.35 0.64 0.36
Q7_10 0.15 -0.11 0.80 -0.14 0.08 -0.04 -0.05 0.02 0.61 0.29 0.68 0.32
Q7_11 0.15 0.10 0.46 -0.21 0.02 0.24 0.07 -0.22 0.47 0.24 0.67 0.33
Q7_12 0.02 0.13 0.16 0.23 0.04 0.20 0.52 -0.11 0.60 0.45 0.57 0.43
Q7_13 -0.08 -0.09 -0.06 0.43 0.00 0.17 0.68 0.09 0.53 0.45 0.54 0.46
Q7_14 -0.07 0.16 -0.10 0.09 -0.04 0.50 0.29 0.19 0.47 0.50 0.49 0.51
Q7_15 0.04 -0.12 0.76 -0.03 -0.02 -0.02 -0.01 0.12 0.48 0.34 0.59 0.41
PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8
SS loadings 7.30 4.95 4.28 3.42 3.84 4.02 1.12 1.06
Proportion Var 0.14 0.10 0.08 0.07 0.08 0.08 0.02 0.02
Cumulative Var 0.14 0.24 0.33 0.39 0.47 0.55 0.57 0.59
Proportion Explained 0.24 0.17 0.14 0.11 0.13 0.13 0.04 0.04
Cumulative Proportion 0.24 0.41 0.55 0.67 0.79 0.93 0.96 1.00
Standardized loadings (pattern matrix)
item PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8 h2 u2
Q4_1 1 0.76 -0.02 0.29 -0.25 -0.04 -0.13 0.07 0.18 0.65 0.35
Q4_2 2 0.77 -0.03 0.16 -0.06 -0.10 -0.06 -0.01 0.14 0.65 0.35
Q4_3 3 0.80 0.06 0.20 -0.28 -0.01 -0.07 0.10 0.06 0.65 0.35
Q4_4 4 0.68 -0.03 0.21 -0.31 -0.02 0.21 -0.07 0.14 0.67 0.33
Q4_5 5 0.49 -0.01 0.21 0.07 -0.02 -0.07 -0.07 0.17 0.54 0.46
Q4_6 6 0.20 0.02 0.18 -0.03 0.10 0.11 0.05 0.51 0.63 0.37
Q4_7 7 0.10 0.09 -0.08 -0.02 0.07 0.40 -0.03 0.38 0.46 0.54
Q4_8 8 0.82 -0.06 -0.04 0.03 0.04 -0.04 -0.05 0.02 0.62 0.38
Q4_9 9 0.73 0.02 -0.14 0.08 0.00 0.11 0.01 -0.18 0.54 0.46
Q4_10 10 0.66 -0.04 0.00 -0.17 0.05 0.23 -0.02 0.13 0.56 0.44
Q4_11 11 0.63 -0.05 0.00 0.20 -0.01 0.07 0.04 0.04 0.61 0.39
Q4_12 12 0.45 -0.13 -0.29 0.52 0.17 0.10 -0.06 0.01 0.58 0.42
Q4_13 13 0.52 -0.03 -0.18 0.43 0.16 -0.17 0.01 -0.10 0.53 0.47
Q4_14 14 0.43 0.01 -0.09 0.44 0.03 -0.03 0.14 0.06 0.55 0.45
Q4_15 15 0.66 0.12 -0.04 0.21 -0.01 -0.09 0.01 -0.06 0.64 0.36
Q4_16 16 0.56 0.22 -0.02 -0.02 0.12 -0.14 -0.02 0.08 0.53 0.47
Q4_17 17 0.66 -0.11 -0.03 0.11 0.06 0.06 -0.07 -0.01 0.51 0.49
Q4_18 18 0.82 0.00 0.05 0.05 -0.11 -0.02 -0.10 0.07 0.74 0.26
Q4_19 19 0.68 0.05 -0.11 0.20 -0.05 0.12 -0.04 -0.07 0.65 0.35
Q5_1 20 0.19 0.33 0.23 0.28 -0.20 -0.11 0.05 -0.04 0.51 0.49
Q5_2 21 -0.01 0.53 -0.03 0.16 -0.02 0.07 -0.08 0.14 0.54 0.46
Q5_3 22 0.05 0.63 0.07 0.30 -0.11 -0.22 -0.03 0.02 0.62 0.38
Q5_4 23 0.01 0.99 -0.14 -0.35 0.06 0.13 0.02 0.04 0.67 0.33
Q5_5 24 0.03 1.11 -0.13 -0.52 0.06 0.10 0.05 -0.10 0.70 0.30
Q5_6 25 0.06 0.80 0.03 -0.07 -0.02 0.04 -0.07 -0.03 0.66 0.34
Q5_7 26 -0.02 0.33 -0.07 0.27 0.19 0.13 -0.07 -0.05 0.48 0.52
Q5_8 27 -0.08 0.70 0.08 0.03 0.00 0.07 -0.07 0.01 0.61 0.39
Q5_9 28 0.03 0.38 0.30 0.20 0.01 -0.12 -0.06 0.08 0.63 0.37
Q5_10 29 0.09 0.33 0.10 0.13 -0.02 0.10 -0.17 -0.02 0.43 0.57
Q5_11 30 0.14 0.20 0.36 0.14 0.03 -0.09 0.22 0.01 0.51 0.49
Q5_12 31 -0.01 0.35 0.20 0.05 0.04 0.14 0.00 0.05 0.47 0.53
Q6_1 32 0.00 0.02 0.06 0.10 0.76 -0.25 0.16 0.05 0.61 0.39
Q6_2 33 -0.05 0.06 0.02 -0.13 0.94 -0.07 0.07 0.12 0.76 0.24
Q6_3 34 -0.04 0.11 -0.10 -0.11 0.93 -0.02 0.03 0.08 0.72 0.28
Q6_4 35 0.00 0.09 -0.06 -0.19 0.88 0.02 -0.06 0.01 0.63 0.37
Q6_5 36 0.00 0.00 0.26 0.25 0.20 -0.09 0.20 -0.07 0.31 0.69
Q6_6 37 0.03 -0.08 0.20 0.09 0.60 0.04 -0.10 -0.05 0.65 0.35
Q6_7 38 0.12 -0.12 0.05 0.21 0.59 0.07 -0.06 -0.06 0.69 0.31
Q6_8 39 -0.01 -0.17 0.16 0.20 0.61 0.06 -0.09 -0.07 0.69 0.31
Q6_9 40 0.04 -0.25 -0.03 0.82 -0.05 0.14 0.27 -0.03 0.48 0.52
Q6_10 41 -0.04 -0.07 0.26 0.44 0.01 0.07 0.15 0.08 0.41 0.59
Q6_11 42 -0.07 -0.06 0.10 0.46 0.10 0.28 0.01 0.24 0.62 0.38
Q7_1 43 -0.03 0.10 0.64 0.06 0.09 0.01 0.05 0.04 0.66 0.34
Q7_2 44 0.02 0.14 0.27 0.02 0.05 0.38 -0.05 -0.05 0.55 0.45
Q7_3 45 -0.06 0.09 -0.18 0.10 0.03 0.78 0.10 0.13 0.66 0.34
Q7_4 46 -0.10 -0.11 0.51 -0.17 0.05 0.59 -0.17 0.08 0.63 0.37
Q7_5 47 0.07 -0.12 0.36 0.14 -0.06 0.52 0.01 -0.02 0.66 0.34
Q7_6 48 0.11 0.00 -0.01 0.24 0.07 0.50 0.08 -0.06 0.57 0.43
Q7_7 49 0.09 0.15 -0.22 0.16 -0.10 0.71 0.18 0.05 0.68 0.32
Q7_8 50 0.09 0.01 0.23 0.14 -0.12 0.52 -0.03 -0.18 0.58 0.42
Q7_9 51 0.08 -0.03 0.84 -0.06 -0.02 -0.03 0.03 0.02 0.64 0.36
Q7_10 52 0.15 -0.11 0.85 -0.14 0.08 -0.05 -0.06 0.02 0.68 0.32
Q7_11 53 0.18 0.12 0.55 -0.25 0.03 0.29 0.08 -0.27 0.67 0.33
Q7_12 54 0.02 0.13 0.15 0.22 0.04 0.20 0.51 -0.11 0.57 0.43
Q7_13 55 -0.08 -0.09 -0.06 0.44 0.00 0.17 0.69 0.09 0.54 0.46
Q7_14 56 -0.08 0.16 -0.10 0.09 -0.04 0.50 0.29 0.19 0.49 0.51
Q7_15 57 0.05 -0.13 0.83 -0.03 -0.03 -0.03 -0.01 0.13 0.59 0.41
PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8
SS loadings 8.20 5.56 4.80 3.84 4.31 4.52 1.25 1.19
Proportion Var 0.14 0.10 0.08 0.07 0.08 0.08 0.02 0.02
Cumulative Var 0.14 0.24 0.33 0.39 0.47 0.55 0.57 0.59
Cum. factor Var 0.24 0.41 0.55 0.67 0.79 0.93 0.96 1.00
With factor correlations of
PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8
PA1 1.00 0.61 0.71 0.63 0.53 0.51 -0.06 0.25
PA4 0.61 1.00 0.71 0.68 0.48 0.57 -0.06 0.40
PA5 0.71 0.71 1.00 0.74 0.62 0.61 -0.07 0.24
PA6 0.63 0.68 0.74 1.00 0.66 0.48 -0.25 0.25
PA3 0.53 0.48 0.62 0.66 1.00 0.37 -0.22 0.18
PA2 0.51 0.57 0.61 0.48 0.37 1.00 0.12 0.16
PA7 -0.06 -0.06 -0.07 -0.25 -0.22 0.12 1.00 -0.05
PA8 0.25 0.40 0.24 0.25 0.18 0.16 -0.05 1.00
Mean item complexity = 1.9
Test of the hypothesis that 8 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1168 and the objective function was 7.45
The root mean square of the residuals (RMSR) is 0.02
The df corrected root mean square of the residuals is 0.03
Fit based upon off diagonal values = 1
Measures of factor score adequacy
PA1 PA4 PA5 PA6 PA3 PA2
Correlation of (regression) scores with factors 0.98 0.97 0.97 0.95 0.97 0.95
Multiple R square of scores with factors 0.96 0.94 0.93 0.90 0.94 0.91
Minimum correlation of possible factor scores 0.91 0.87 0.87 0.80 0.88 0.82
PA7 PA8
Correlation of (regression) scores with factors 0.86 0.84
Multiple R square of scores with factors 0.73 0.71
Minimum correlation of possible factor scores 0.46 0.42print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8
Q4_1 0.661
Q4_2 0.635
Q4_3 0.670
Q4_4 0.558
Q4_5 0.435
Q4_6 0.502
Q4_7 0.375 0.357
Q4_8 0.760
Q4_9 0.731
Q4_10 0.522
Q4_11 0.604
Q4_12 0.469 -0.303 0.546
Q4_13 0.458 0.381
Q4_14 0.388 0.395
Q4_15 0.603
Q4_16 0.531
Q4_17 0.634
Q4_18 0.672
Q4_19 0.659
Q5_1 0.315
Q5_2 0.550
Q5_3 0.655 0.309
Q5_4 1.092 -0.387
Q5_5 1.151 -0.541
Q5_6 0.724
Q5_7 0.308
Q5_8 0.737
Q5_9 0.374 0.303
Q5_10
Q5_11 0.344
Q5_12 0.336
Q6_1 0.637
Q6_2 0.860
Q6_3 0.860
Q6_4 0.815
Q6_5
Q6_6 0.488
Q6_7 0.513
Q6_8 0.554
Q6_9 -0.303 1.002 0.328
Q6_10 0.404
Q6_11 0.443
Q7_1 0.578
Q7_2 0.324
Q7_3 0.790
Q7_4 0.502 0.575
Q7_5 0.341 0.489
Q7_6 0.458
Q7_7 0.721
Q7_8 0.452
Q7_9 0.836
Q7_10 0.804
Q7_11 0.458
Q7_12 0.522
Q7_13 0.434 0.685
Q7_14 0.498
Q7_15 0.756
PA1 PA4 PA5 PA6 PA3 PA2 PA7 PA8
SS loadings 6.305 5.305 3.971 3.734 3.659 3.287 1.281 0.844
Proportion Var 0.111 0.093 0.070 0.066 0.064 0.058 0.022 0.015
Cumulative Var 0.111 0.204 0.273 0.339 0.403 0.461 0.483 0.498Nine factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 9, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 9, rotate = "Promax", fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA3 PA4 PA2 PA9 PA6 PA5 PA1 PA7 PA8 h2 u2 com
Q4_1 0.95 -0.03 -0.01 -0.13 -0.17 0.20 -0.07 0.07 0.15 0.73 0.27 1.3
Q4_2 0.81 -0.02 -0.12 -0.14 0.01 0.21 0.17 -0.04 0.14 0.74 0.26 1.4
Q4_3 0.98 0.03 0.03 -0.23 -0.10 0.11 -0.09 0.11 0.02 0.74 0.26 1.2
Q4_4 0.86 -0.04 -0.01 -0.12 0.18 0.14 -0.14 -0.08 0.13 0.78 0.22 1.3
Q4_5 0.48 0.04 -0.05 0.00 0.02 0.27 0.23 -0.11 0.18 0.64 0.36 2.6
Q4_6 0.34 0.09 0.10 0.11 0.13 0.19 -0.08 0.04 0.50 0.68 0.32 2.7
Q4_7 0.16 0.13 0.06 0.29 0.33 -0.08 -0.20 -0.03 0.40 0.51 0.49 4.3
Q4_8 0.85 -0.08 0.02 -0.01 -0.02 -0.04 0.13 -0.05 0.03 0.68 0.32 1.1
Q4_9 0.65 0.01 0.05 -0.26 0.20 -0.07 0.25 0.04 -0.20 0.60 0.40 2.2
Q4_10 0.80 -0.05 0.07 0.01 0.20 -0.08 -0.11 -0.04 0.09 0.68 0.32 1.3
Q4_11 0.63 -0.04 -0.02 0.24 0.05 -0.05 0.07 0.05 0.02 0.67 0.33 1.4
Q4_12 0.27 -0.12 0.15 0.19 0.25 -0.17 0.52 -0.08 0.05 0.75 0.25 3.1
Q4_13 0.30 -0.03 0.14 -0.09 0.01 0.03 0.74 -0.02 -0.12 0.89 0.11 1.5
Q4_14 0.39 0.02 0.07 0.47 -0.11 -0.12 0.13 0.19 0.06 0.64 0.36 2.9
Q4_15 0.65 0.14 -0.02 0.35 -0.16 -0.15 0.02 0.03 -0.09 0.73 0.27 2.0
Q4_16 0.65 0.24 0.15 0.26 -0.28 -0.15 -0.13 0.02 0.06 0.65 0.35 2.5
Q4_17 0.66 -0.09 0.02 0.20 0.03 -0.08 0.05 -0.07 -0.02 0.58 0.42 1.3
Q4_18 0.84 0.05 -0.14 0.08 -0.01 0.03 0.13 -0.12 0.05 0.84 0.16 1.2
Q4_19 0.60 0.06 -0.01 0.03 0.16 -0.08 0.22 0.00 -0.08 0.70 0.30 1.5
Q5_1 0.14 0.39 -0.17 0.15 -0.07 0.21 0.17 0.05 -0.07 0.57 0.43 3.4
Q5_2 -0.07 0.59 0.02 0.02 0.11 0.06 0.21 -0.07 0.15 0.59 0.41 1.6
Q5_3 -0.06 0.70 -0.06 0.10 -0.16 0.13 0.25 -0.02 0.01 0.70 0.30 1.5
Q5_4 0.01 0.92 0.01 -0.25 0.18 -0.09 -0.05 0.00 0.09 0.70 0.30 1.3
Q5_5 0.05 0.98 0.04 -0.40 0.15 -0.10 -0.11 0.03 -0.04 0.67 0.33 1.4
Q5_6 0.09 0.81 -0.01 0.10 -0.02 -0.05 -0.09 -0.04 -0.02 0.72 0.28 1.1
Q5_7 -0.07 0.33 0.19 0.54 0.06 -0.20 -0.07 -0.06 -0.04 0.58 0.42 2.5
Q5_8 -0.11 0.74 0.00 0.19 0.06 0.00 -0.04 -0.06 0.01 0.67 0.33 1.2
Q5_9 0.05 0.46 0.01 0.45 -0.18 0.16 -0.06 -0.06 0.06 0.71 0.29 2.7
Q5_10 0.07 0.37 -0.04 0.34 0.04 0.01 -0.01 -0.17 -0.06 0.52 0.48 2.6
Q5_11 0.14 0.25 0.05 0.12 -0.06 0.26 0.01 0.23 -0.01 0.56 0.44 4.2
Q5_12 -0.04 0.41 0.07 0.06 0.15 0.18 0.03 0.01 0.03 0.51 0.49 1.8
Q6_1 -0.02 0.00 0.81 0.00 -0.25 0.14 0.15 0.19 0.02 0.78 0.22 1.5
Q6_2 -0.05 0.07 0.98 -0.14 -0.03 0.05 0.05 0.07 0.11 0.85 0.15 1.1
Q6_3 -0.02 0.09 0.93 -0.08 0.04 -0.08 0.04 -0.01 0.07 0.81 0.19 1.1
Q6_4 0.00 0.07 0.92 -0.11 0.02 -0.08 -0.01 -0.04 0.00 0.71 0.29 1.1
Q6_5 -0.04 -0.02 0.21 0.22 -0.05 0.23 0.08 0.16 -0.06 0.36 0.64 4.4
Q6_6 -0.03 -0.11 0.53 0.25 0.07 0.21 0.05 -0.16 -0.02 0.77 0.23 2.2
Q6_7 0.10 -0.14 0.56 0.39 0.04 -0.02 0.02 -0.06 -0.03 0.80 0.20 2.1
Q6_8 -0.06 -0.16 0.60 0.41 0.03 0.07 -0.02 -0.08 -0.05 0.79 0.21 2.1
Q6_9 -0.07 -0.25 -0.07 0.83 0.10 -0.06 0.12 0.29 0.00 0.57 0.43 1.6
Q6_10 -0.03 -0.08 -0.05 0.83 -0.07 0.15 -0.15 0.18 0.14 0.53 0.47 1.4
Q6_11 -0.07 -0.03 0.07 0.70 0.20 0.08 -0.02 0.02 0.28 0.70 0.30 1.6
Q7_1 -0.04 0.14 0.12 0.03 0.09 0.60 0.08 0.05 0.03 0.75 0.25 1.3
Q7_2 -0.01 0.12 0.07 0.01 0.44 0.24 0.06 -0.03 -0.03 0.60 0.40 1.9
Q7_3 -0.10 0.10 0.05 -0.04 0.88 -0.13 0.07 0.13 0.15 0.72 0.28 1.2
Q7_4 -0.08 -0.04 0.06 -0.08 0.64 0.45 -0.07 -0.13 0.06 0.67 0.33 2.1
Q7_5 0.07 -0.09 -0.05 0.17 0.52 0.27 0.00 0.07 -0.03 0.70 0.30 1.9
Q7_6 0.06 0.03 0.11 0.06 0.52 -0.01 0.12 0.15 -0.07 0.61 0.39 1.5
Q7_7 0.04 0.14 -0.15 0.02 0.79 -0.15 0.11 0.25 0.10 0.79 0.21 1.5
Q7_8 0.03 0.03 -0.14 0.18 0.51 0.16 -0.02 0.02 -0.19 0.63 0.37 2.0
Q7_9 0.12 0.02 0.01 0.00 0.01 0.73 -0.01 0.06 -0.01 0.71 0.29 1.1
Q7_10 0.20 -0.05 0.07 -0.11 0.05 0.79 0.02 -0.08 0.02 0.78 0.22 1.2
Q7_11 0.25 0.06 0.03 -0.02 0.26 0.36 -0.23 0.12 -0.31 0.78 0.22 4.9
Q7_12 -0.01 0.05 0.04 0.14 0.22 0.08 0.00 0.59 -0.14 0.67 0.33 1.6
Q7_13 -0.08 -0.13 0.00 0.29 0.21 -0.06 -0.07 0.70 0.07 0.58 0.42 1.7
Q7_14 -0.03 0.15 -0.07 0.08 0.52 -0.08 -0.11 0.34 0.16 0.52 0.48 2.4
Q7_15 0.11 -0.08 -0.03 0.13 0.00 0.75 -0.06 -0.02 0.14 0.67 0.33 1.2
PA3 PA4 PA2 PA9 PA6 PA5 PA1 PA7 PA8
SS loadings 9.71 5.47 5.15 4.87 4.84 4.27 1.94 1.67 0.68
Proportion Var 0.17 0.10 0.09 0.09 0.08 0.07 0.03 0.03 0.01
Cumulative Var 0.17 0.27 0.36 0.44 0.53 0.60 0.64 0.67 0.68
Proportion Explained 0.25 0.14 0.13 0.13 0.13 0.11 0.05 0.04 0.02
Cumulative Proportion 0.25 0.39 0.53 0.65 0.78 0.89 0.94 0.98 1.00
With factor correlations of
PA3 PA4 PA2 PA9 PA6 PA5 PA1 PA7 PA8
PA3 1.00 0.64 0.54 0.72 0.58 0.68 0.43 0.12 -0.21
PA4 0.64 1.00 0.47 0.67 0.58 0.64 0.37 0.21 -0.04
PA2 0.54 0.47 1.00 0.67 0.41 0.58 0.41 -0.05 -0.16
PA9 0.72 0.67 0.67 1.00 0.64 0.73 0.52 0.06 -0.26
PA6 0.58 0.58 0.41 0.64 1.00 0.57 0.19 0.21 -0.19
PA5 0.68 0.64 0.58 0.73 0.57 1.00 0.40 0.16 -0.20
PA1 0.43 0.37 0.41 0.52 0.19 0.40 1.00 0.06 -0.02
PA7 0.12 0.21 -0.05 0.06 0.21 0.16 0.06 1.00 0.05
PA8 -0.21 -0.04 -0.16 -0.26 -0.19 -0.20 -0.02 0.05 1.00
Mean item complexity = 1.9
Test of the hypothesis that 9 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1119 and the objective function was 35.98
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.03
Fit based upon off diagonal values = 1print(fit$loadings, cutoff=0.3)
Loadings:
PA3 PA4 PA2 PA9 PA6 PA5 PA1 PA7 PA8
Q4_1 0.951
Q4_2 0.812
Q4_3 0.977
Q4_4 0.865
Q4_5 0.476
Q4_6 0.339 0.500
Q4_7 0.330 0.397
Q4_8 0.852
Q4_9 0.651
Q4_10 0.796
Q4_11 0.632
Q4_12 0.518
Q4_13 0.737
Q4_14 0.394 0.468
Q4_15 0.649 0.347
Q4_16 0.651
Q4_17 0.657
Q4_18 0.841
Q4_19 0.599
Q5_1 0.389
Q5_2 0.587
Q5_3 0.698
Q5_4 0.925
Q5_5 0.977 -0.398
Q5_6 0.811
Q5_7 0.333 0.537
Q5_8 0.744
Q5_9 0.464 0.451
Q5_10 0.367 0.344
Q5_11
Q5_12 0.410
Q6_1 0.811
Q6_2 0.978
Q6_3 0.935
Q6_4 0.919
Q6_5
Q6_6 0.533
Q6_7 0.563 0.393
Q6_8 0.603 0.409
Q6_9 0.834
Q6_10 0.827
Q6_11 0.698
Q7_1 0.598
Q7_2 0.444
Q7_3 0.878
Q7_4 0.639 0.447
Q7_5 0.520
Q7_6 0.522
Q7_7 0.788
Q7_8 0.507
Q7_9 0.732
Q7_10 0.791
Q7_11 0.362 -0.309
Q7_12 0.585
Q7_13 0.696
Q7_14 0.522 0.336
Q7_15 0.754
PA3 PA4 PA2 PA9 PA6 PA5 PA1 PA7 PA8
SS loadings 8.753 5.119 4.663 4.310 4.018 3.395 1.535 1.527 1.010
Proportion Var 0.154 0.090 0.082 0.076 0.070 0.060 0.027 0.027 0.018
Cumulative Var 0.154 0.243 0.325 0.401 0.471 0.531 0.558 0.585 0.602fit <- psych::fa(r = cov.dat, nfactors = 9, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 9, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8 h2 u2 H2 U2
Q4_1 0.69 0.02 -0.15 0.21 -0.04 -0.14 0.05 -0.09 0.16 0.49 0.26 0.65 0.35
Q4_2 0.66 -0.03 -0.10 0.14 -0.07 -0.04 -0.01 0.02 0.11 0.43 0.24 0.65 0.35
Q4_3 0.70 0.10 -0.19 0.13 -0.02 -0.08 0.08 -0.11 0.05 0.45 0.25 0.65 0.35
Q4_4 0.58 0.04 -0.10 0.10 -0.04 0.11 -0.06 -0.18 0.12 0.46 0.21 0.69 0.31
Q4_5 0.46 -0.05 -0.01 0.20 0.00 -0.03 -0.06 0.10 0.15 0.42 0.35 0.54 0.46
Q4_6 0.22 -0.02 -0.03 0.18 0.11 0.13 0.04 0.06 0.49 0.59 0.37 0.62 0.38
Q4_7 0.10 0.07 0.08 -0.11 0.05 0.34 -0.03 -0.03 0.36 0.40 0.48 0.46 0.54
Q4_8 0.78 -0.04 -0.03 -0.03 0.03 -0.04 -0.04 0.01 0.02 0.53 0.33 0.61 0.39
Q4_9 0.76 0.00 -0.23 -0.02 0.04 0.21 0.02 0.15 -0.20 0.59 0.42 0.59 0.41
Q4_10 0.55 0.05 0.04 -0.09 0.01 0.10 -0.02 -0.19 0.10 0.38 0.25 0.60 0.40
Q4_11 0.63 -0.03 0.22 -0.04 -0.03 0.03 0.03 -0.02 0.04 0.56 0.35 0.61 0.39
Q4_12 0.48 -0.20 0.32 -0.23 0.19 0.16 -0.06 0.23 0.01 0.66 0.45 0.60 0.40
Q4_13 0.47 -0.10 0.12 -0.06 0.17 -0.07 0.01 0.24 -0.09 0.43 0.34 0.56 0.44
Q4_14 0.40 -0.01 0.37 -0.09 0.02 -0.04 0.12 0.09 0.05 0.44 0.36 0.55 0.45
Q4_15 0.62 0.11 0.24 -0.08 -0.03 -0.12 0.01 0.03 -0.06 0.54 0.30 0.64 0.36
Q4_16 0.55 0.20 0.05 -0.06 0.10 -0.16 -0.02 0.02 0.08 0.47 0.41 0.54 0.46
Q4_17 0.65 -0.09 0.05 -0.03 0.05 0.06 -0.06 0.01 -0.01 0.47 0.46 0.51 0.49
Q4_18 0.70 0.01 0.06 0.01 -0.10 -0.04 -0.08 0.00 0.06 0.50 0.18 0.74 0.26
Q4_19 0.68 0.00 -0.01 -0.03 -0.02 0.18 -0.03 0.16 -0.08 0.63 0.31 0.67 0.33
Q5_1 0.19 0.20 0.17 0.26 -0.17 -0.06 0.05 0.25 -0.04 0.46 0.44 0.51 0.49
Q5_2 -0.01 0.35 -0.10 0.11 0.03 0.21 -0.09 0.45 0.16 0.66 0.41 0.62 0.38
Q5_3 0.04 0.43 0.03 0.23 -0.07 -0.10 -0.02 0.56 0.02 0.77 0.32 0.71 0.29
Q5_4 0.01 1.01 -0.21 -0.17 0.06 0.11 0.03 0.08 0.04 0.82 0.40 0.67 0.33
Q5_5 0.04 1.12 -0.29 -0.18 0.05 0.04 0.05 -0.02 -0.10 0.77 0.30 0.72 0.28
Q5_6 0.06 0.63 0.04 0.01 -0.02 0.02 -0.06 0.14 -0.02 0.54 0.28 0.66 0.34
Q5_7 -0.03 0.32 0.60 -0.23 0.11 -0.01 -0.07 -0.06 -0.05 0.50 0.39 0.56 0.44
Q5_8 -0.09 0.64 0.21 0.04 -0.02 0.03 -0.07 0.14 0.01 0.68 0.43 0.61 0.39
Q5_9 0.03 0.31 0.43 0.20 -0.03 -0.20 -0.06 0.09 0.08 0.65 0.34 0.65 0.35
Q5_10 0.08 0.24 0.27 0.01 -0.05 0.02 -0.14 0.03 -0.02 0.31 0.39 0.45 0.55
Q5_11 0.14 0.16 0.13 0.34 0.03 -0.07 0.21 0.08 0.00 0.46 0.45 0.51 0.49
Q5_12 0.00 0.25 -0.01 0.23 0.06 0.17 0.00 0.16 0.05 0.43 0.47 0.48 0.52
Q6_1 0.00 -0.01 -0.02 0.10 0.65 -0.16 0.13 0.09 0.04 0.45 0.26 0.63 0.37
Q6_2 -0.05 0.05 -0.19 0.07 0.87 -0.01 0.06 0.03 0.11 0.65 0.19 0.78 0.22
Q6_3 -0.04 0.12 -0.05 -0.09 0.84 -0.02 0.03 -0.05 0.07 0.61 0.25 0.71 0.29
Q6_4 -0.01 0.08 -0.23 0.00 0.82 0.06 -0.06 0.01 0.01 0.55 0.31 0.64 0.36
Q6_5 0.00 0.02 0.39 0.19 0.18 -0.14 0.21 -0.05 -0.07 0.34 0.72 0.32 0.68
Q6_6 0.02 -0.05 0.16 0.12 0.46 0.00 -0.08 -0.05 -0.04 0.44 0.24 0.65 0.35
Q6_7 0.11 -0.07 0.30 -0.01 0.48 0.01 -0.05 -0.07 -0.05 0.54 0.23 0.70 0.30
Q6_8 -0.01 -0.13 0.29 0.09 0.52 0.01 -0.08 -0.06 -0.06 0.57 0.25 0.70 0.30
Q6_9 0.05 -0.30 1.10 -0.13 -0.10 0.09 0.33 0.04 -0.03 0.75 0.74 0.50 0.50
Q6_10 -0.04 -0.06 0.62 0.13 -0.03 -0.03 0.14 -0.05 0.08 0.38 0.46 0.45 0.55
Q6_11 -0.07 -0.09 0.61 0.02 0.06 0.20 0.01 0.02 0.24 0.60 0.34 0.64 0.36
Q7_1 -0.02 0.03 0.04 0.58 0.09 0.04 0.05 0.09 0.03 0.54 0.27 0.66 0.34
Q7_2 0.01 0.11 0.09 0.20 0.04 0.30 -0.04 -0.02 -0.04 0.40 0.33 0.55 0.45
Q7_3 -0.06 0.08 0.04 -0.14 0.05 0.80 0.10 -0.01 0.14 0.68 0.35 0.66 0.34
Q7_4 -0.10 -0.11 -0.17 0.52 0.05 0.58 -0.17 -0.05 0.08 0.61 0.35 0.64 0.36
Q7_5 0.07 -0.11 0.12 0.34 -0.06 0.48 0.01 -0.02 -0.02 0.60 0.30 0.66 0.34
Q7_6 0.10 -0.01 0.13 0.04 0.07 0.48 0.08 0.05 -0.05 0.48 0.36 0.57 0.43
Q7_7 0.09 0.13 0.01 -0.14 -0.08 0.77 0.18 0.05 0.06 0.72 0.31 0.70 0.30
Q7_8 0.08 0.02 0.17 0.18 -0.12 0.41 -0.02 -0.05 -0.15 0.44 0.33 0.58 0.42
Q7_9 0.08 -0.09 -0.13 0.87 0.00 0.02 0.03 0.08 0.02 0.65 0.33 0.66 0.34
Q7_10 0.16 -0.14 -0.18 0.82 0.09 -0.01 -0.05 0.03 0.02 0.61 0.28 0.69 0.31
Q7_11 0.15 0.16 -0.07 0.40 0.00 0.19 0.07 -0.17 -0.22 0.47 0.23 0.67 0.33
Q7_12 0.01 0.16 0.22 0.16 0.04 0.21 0.51 -0.05 -0.11 0.60 0.44 0.57 0.43
Q7_13 -0.08 -0.05 0.38 -0.05 0.01 0.19 0.65 -0.04 0.09 0.51 0.47 0.52 0.48
Q7_14 -0.07 0.15 0.07 -0.08 -0.02 0.51 0.28 -0.01 0.19 0.47 0.50 0.49 0.51
Q7_15 0.06 -0.15 -0.01 0.73 -0.02 -0.01 -0.01 0.02 0.12 0.48 0.34 0.59 0.41
PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8
SS loadings 7.63 4.12 3.79 4.07 3.73 3.91 1.06 1.32 1.03
Proportion Var 0.15 0.08 0.07 0.08 0.07 0.08 0.02 0.03 0.02
Cumulative Var 0.15 0.23 0.31 0.39 0.46 0.54 0.56 0.58 0.60
Proportion Explained 0.25 0.13 0.12 0.13 0.12 0.13 0.03 0.04 0.03
Cumulative Proportion 0.25 0.38 0.51 0.64 0.76 0.89 0.92 0.97 1.00
Standardized loadings (pattern matrix)
item PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8 h2 u2
Q4_1 1 0.80 0.02 -0.18 0.24 -0.05 -0.16 0.06 -0.11 0.18 0.65 0.35
Q4_2 2 0.81 -0.04 -0.12 0.17 -0.09 -0.05 -0.01 0.03 0.13 0.65 0.35
Q4_3 3 0.83 0.11 -0.22 0.16 -0.02 -0.10 0.10 -0.13 0.06 0.65 0.35
Q4_4 4 0.72 0.05 -0.12 0.12 -0.05 0.13 -0.07 -0.22 0.14 0.69 0.31
Q4_5 5 0.52 -0.05 -0.01 0.23 0.00 -0.04 -0.07 0.12 0.17 0.54 0.46
Q4_6 6 0.22 -0.02 -0.04 0.18 0.12 0.13 0.04 0.06 0.50 0.62 0.38
Q4_7 7 0.11 0.08 0.08 -0.12 0.06 0.36 -0.03 -0.04 0.38 0.46 0.54
Q4_8 8 0.85 -0.05 -0.03 -0.04 0.03 -0.04 -0.05 0.01 0.02 0.61 0.39
Q4_9 9 0.76 0.00 -0.23 -0.02 0.04 0.21 0.02 0.14 -0.20 0.59 0.41
Q4_10 10 0.70 0.06 0.05 -0.11 0.01 0.13 -0.03 -0.24 0.13 0.60 0.40
Q4_11 11 0.66 -0.03 0.23 -0.04 -0.03 0.03 0.03 -0.02 0.04 0.61 0.39
Q4_12 12 0.46 -0.19 0.30 -0.22 0.18 0.15 -0.06 0.22 0.01 0.60 0.40
Q4_13 13 0.54 -0.11 0.13 -0.07 0.19 -0.08 0.01 0.28 -0.11 0.56 0.44
Q4_14 14 0.45 -0.02 0.41 -0.10 0.02 -0.04 0.14 0.10 0.06 0.55 0.45
Q4_15 15 0.68 0.12 0.26 -0.09 -0.03 -0.13 0.01 0.03 -0.06 0.64 0.36
Q4_16 16 0.59 0.21 0.06 -0.06 0.10 -0.17 -0.02 0.02 0.08 0.54 0.46
Q4_17 17 0.68 -0.10 0.05 -0.03 0.05 0.06 -0.07 0.01 -0.01 0.51 0.49
Q4_18 18 0.85 0.01 0.07 0.01 -0.12 -0.05 -0.10 0.00 0.07 0.74 0.26
Q4_19 19 0.70 0.00 -0.01 -0.03 -0.02 0.18 -0.03 0.16 -0.08 0.67 0.33
Q5_1 20 0.20 0.21 0.18 0.28 -0.18 -0.06 0.05 0.26 -0.04 0.51 0.49
Q5_2 21 -0.01 0.34 -0.09 0.11 0.03 0.20 -0.09 0.43 0.16 0.62 0.38
Q5_3 22 0.04 0.41 0.03 0.22 -0.07 -0.10 -0.02 0.53 0.02 0.71 0.29
Q5_4 23 0.01 0.91 -0.19 -0.15 0.06 0.10 0.02 0.07 0.04 0.67 0.33
Q5_5 24 0.03 1.09 -0.28 -0.18 0.05 0.04 0.05 -0.02 -0.10 0.72 0.28
Q5_6 25 0.06 0.69 0.04 0.01 -0.03 0.02 -0.06 0.16 -0.02 0.66 0.34
Q5_7 26 -0.03 0.34 0.64 -0.24 0.12 -0.01 -0.08 -0.06 -0.06 0.56 0.44
Q5_8 27 -0.08 0.60 0.20 0.03 -0.02 0.03 -0.07 0.14 0.01 0.61 0.39
Q5_9 28 0.03 0.31 0.43 0.21 -0.03 -0.20 -0.06 0.09 0.08 0.65 0.35
Q5_10 29 0.09 0.29 0.33 0.02 -0.06 0.03 -0.17 0.04 -0.02 0.45 0.55
Q5_11 30 0.14 0.16 0.13 0.36 0.04 -0.08 0.22 0.08 0.00 0.51 0.49
Q5_12 31 0.00 0.26 -0.01 0.24 0.06 0.18 0.00 0.17 0.05 0.48 0.52
Q6_1 32 0.00 -0.01 -0.03 0.12 0.77 -0.18 0.16 0.11 0.05 0.63 0.37
Q6_2 33 -0.05 0.05 -0.21 0.08 0.96 -0.01 0.07 0.03 0.12 0.78 0.22
Q6_3 34 -0.04 0.13 -0.06 -0.09 0.91 -0.02 0.03 -0.05 0.08 0.71 0.29
Q6_4 35 -0.01 0.09 -0.25 0.00 0.88 0.07 -0.06 0.01 0.01 0.64 0.36
Q6_5 36 0.00 0.02 0.38 0.19 0.17 -0.14 0.20 -0.05 -0.07 0.32 0.68
Q6_6 37 0.03 -0.06 0.20 0.15 0.56 -0.01 -0.10 -0.06 -0.05 0.65 0.35
Q6_7 38 0.13 -0.08 0.35 -0.02 0.54 0.01 -0.06 -0.08 -0.06 0.70 0.30
Q6_8 39 -0.01 -0.14 0.32 0.10 0.57 0.01 -0.09 -0.07 -0.07 0.70 0.30
Q6_9 40 0.04 -0.25 0.90 -0.11 -0.09 0.07 0.27 0.03 -0.03 0.50 0.50
Q6_10 41 -0.04 -0.07 0.68 0.14 -0.04 -0.03 0.15 -0.05 0.09 0.45 0.55
Q6_11 42 -0.07 -0.09 0.63 0.02 0.06 0.21 0.01 0.02 0.25 0.64 0.36
Q7_1 43 -0.03 0.04 0.04 0.65 0.10 0.04 0.05 0.10 0.03 0.66 0.34
Q7_2 44 0.02 0.13 0.11 0.24 0.04 0.35 -0.04 -0.02 -0.05 0.55 0.45
Q7_3 45 -0.06 0.08 0.04 -0.14 0.05 0.79 0.10 -0.01 0.14 0.66 0.34
Q7_4 46 -0.10 -0.12 -0.17 0.53 0.06 0.60 -0.17 -0.05 0.08 0.64 0.36
Q7_5 47 0.07 -0.12 0.13 0.36 -0.06 0.51 0.01 -0.03 -0.02 0.66 0.34
Q7_6 48 0.11 -0.02 0.14 0.04 0.08 0.52 0.08 0.05 -0.05 0.57 0.43
Q7_7 49 0.09 0.13 0.01 -0.14 -0.08 0.76 0.18 0.05 0.06 0.70 0.30
Q7_8 50 0.09 0.02 0.20 0.20 -0.13 0.47 -0.02 -0.06 -0.17 0.58 0.42
Q7_9 51 0.08 -0.09 -0.13 0.88 0.00 0.02 0.03 0.08 0.02 0.66 0.34
Q7_10 52 0.17 -0.14 -0.19 0.87 0.10 -0.01 -0.05 0.03 0.02 0.69 0.31
Q7_11 53 0.18 0.19 -0.08 0.48 0.00 0.23 0.08 -0.20 -0.27 0.67 0.33
Q7_12 54 0.01 0.16 0.21 0.15 0.04 0.20 0.50 -0.05 -0.11 0.57 0.43
Q7_13 55 -0.08 -0.05 0.38 -0.06 0.01 0.19 0.66 -0.04 0.09 0.52 0.48
Q7_14 56 -0.07 0.16 0.07 -0.08 -0.02 0.51 0.29 -0.01 0.20 0.49 0.51
Q7_15 57 0.06 -0.16 -0.01 0.81 -0.02 -0.02 -0.01 0.03 0.13 0.59 0.41
PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8
SS loadings 8.56 4.63 4.25 4.57 4.19 4.39 1.19 1.49 1.16
Proportion Var 0.15 0.08 0.07 0.08 0.07 0.08 0.02 0.03 0.02
Cumulative Var 0.15 0.23 0.31 0.39 0.46 0.54 0.56 0.58 0.60
Cum. factor Var 0.25 0.38 0.51 0.64 0.76 0.89 0.92 0.97 1.00
With factor correlations of
PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8
PA1 1.00 0.57 0.70 0.71 0.53 0.53 -0.09 0.27 0.20
PA4 0.57 1.00 0.64 0.69 0.38 0.58 -0.08 0.18 0.39
PA9 0.70 0.64 1.00 0.79 0.67 0.62 -0.21 0.32 0.20
PA6 0.71 0.69 0.79 1.00 0.58 0.61 -0.10 0.20 0.22
PA3 0.53 0.38 0.67 0.58 1.00 0.36 -0.23 0.30 0.10
PA2 0.53 0.58 0.62 0.61 0.36 1.00 0.04 0.13 0.15
PA7 -0.09 -0.08 -0.21 -0.10 -0.23 0.04 1.00 -0.04 -0.03
PA5 0.27 0.18 0.32 0.20 0.30 0.13 -0.04 1.00 0.05
PA8 0.20 0.39 0.20 0.22 0.10 0.15 -0.03 0.05 1.00
Mean item complexity = 1.9
Test of the hypothesis that 9 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1119 and the objective function was 6.84
The root mean square of the residuals (RMSR) is 0.02
The df corrected root mean square of the residuals is 0.03
Fit based upon off diagonal values = 1
Measures of factor score adequacy
PA1 PA4 PA9 PA6 PA3 PA2
Correlation of (regression) scores with factors 0.98 0.96 0.96 0.96 0.97 0.95
Multiple R square of scores with factors 0.96 0.92 0.92 0.93 0.94 0.91
Minimum correlation of possible factor scores 0.92 0.85 0.84 0.86 0.88 0.82
PA7 PA5 PA8
Correlation of (regression) scores with factors 0.85 0.86 0.84
Multiple R square of scores with factors 0.72 0.74 0.71
Minimum correlation of possible factor scores 0.44 0.49 0.42print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8
Q4_1 0.691
Q4_2 0.661
Q4_3 0.695
Q4_4 0.585
Q4_5 0.457
Q4_6 0.492
Q4_7 0.340 0.358
Q4_8 0.785
Q4_9 0.762
Q4_10 0.549
Q4_11 0.626
Q4_12 0.484 0.316
Q4_13 0.473
Q4_14 0.402 0.369
Q4_15 0.623
Q4_16 0.553
Q4_17 0.652
Q4_18 0.699
Q4_19 0.681
Q5_1
Q5_2 0.353 0.446
Q5_3 0.432 0.559
Q5_4 1.010
Q5_5 1.122
Q5_6 0.628
Q5_7 0.319 0.604
Q5_8 0.635
Q5_9 0.306 0.432
Q5_10
Q5_11 0.340
Q5_12
Q6_1 0.653
Q6_2 0.874
Q6_3 0.843
Q6_4 0.818
Q6_5 0.391
Q6_6 0.462
Q6_7 0.304 0.476
Q6_8 0.519
Q6_9 -0.304 1.099 0.331
Q6_10 0.623
Q6_11 0.612
Q7_1 0.585
Q7_2
Q7_3 0.804
Q7_4 0.517 0.584
Q7_5 0.341 0.482
Q7_6 0.481
Q7_7 0.769
Q7_8 0.414
Q7_9 0.870
Q7_10 0.820
Q7_11 0.402
Q7_12 0.511
Q7_13 0.375 0.651
Q7_14 0.507
Q7_15 0.735
PA1 PA4 PA9 PA6 PA3 PA2 PA7 PA5 PA8
SS loadings 6.802 4.244 4.127 3.844 3.559 3.256 1.213 1.024 0.857
Proportion Var 0.119 0.074 0.072 0.067 0.062 0.057 0.021 0.018 0.015
Cumulative Var 0.119 0.194 0.266 0.334 0.396 0.453 0.474 0.492 0.507Ten factor extraction
fit <- psych::fa(r = corr.pc$rho, nfactors = 10, rotate = "Promax", fm = "pa")Warning in fa.stats(r = r, f = f, phi = phi, n.obs = n.obs, np.obs = np.obs, :
The estimated weights for the factor scores are probably incorrect. Try a
different factor score estimation method.fitFactor Analysis using method = pa
Call: psych::fa(r = corr.pc$rho, nfactors = 10, rotate = "Promax",
fm = "pa")
Standardized loadings (pattern matrix) based upon correlation matrix
PA3 PA2 PA6 PA10 PA4 PA5 PA9 PA7 PA1 PA8 h2 u2
Q4_1 0.96 -0.02 -0.18 -0.14 0.00 0.19 -0.05 0.08 -0.04 0.14 0.73 0.267
Q4_2 0.84 -0.12 -0.03 -0.10 0.02 0.19 -0.03 -0.03 0.18 0.12 0.75 0.245
Q4_3 0.99 0.02 -0.12 -0.21 0.07 0.11 -0.10 0.12 -0.05 0.02 0.74 0.257
Q4_4 0.89 -0.02 0.14 -0.06 0.03 0.12 -0.22 -0.07 -0.07 0.11 0.79 0.210
Q4_5 0.50 -0.05 -0.02 0.04 0.05 0.24 0.05 -0.10 0.22 0.17 0.65 0.353
Q4_6 0.33 0.11 0.15 0.04 0.03 0.20 0.13 0.03 -0.09 0.52 0.68 0.318
Q4_7 0.15 0.07 0.36 0.24 0.06 -0.07 0.05 -0.04 -0.20 0.42 0.52 0.482
Q4_8 0.85 0.02 -0.02 -0.01 -0.06 -0.04 0.00 -0.05 0.12 0.03 0.68 0.318
Q4_9 0.62 0.07 0.27 -0.34 -0.04 -0.03 0.17 0.02 0.17 -0.19 0.63 0.367
Q4_10 0.84 0.06 0.14 0.11 0.06 -0.12 -0.29 -0.01 -0.03 0.07 0.72 0.284
Q4_11 0.64 -0.02 0.05 0.24 -0.03 -0.05 0.00 0.05 0.07 0.02 0.67 0.328
Q4_12 0.29 0.14 0.21 0.25 -0.08 -0.20 0.05 -0.06 0.47 0.03 0.76 0.241
Q4_13 0.33 0.13 -0.04 -0.01 0.01 -0.01 0.12 0.00 0.69 -0.15 0.93 0.073
Q4_14 0.37 0.07 -0.06 0.36 -0.06 -0.10 0.24 0.18 0.06 0.07 0.65 0.349
Q4_15 0.63 -0.01 -0.12 0.28 0.04 -0.12 0.22 0.01 -0.04 -0.07 0.75 0.245
Q4_16 0.62 0.17 -0.22 0.16 0.09 -0.11 0.31 0.00 -0.22 0.10 0.72 0.275
Q4_17 0.63 0.03 0.08 0.12 -0.15 -0.05 0.14 -0.09 -0.01 0.01 0.60 0.402
Q4_18 0.86 -0.15 -0.04 0.12 0.06 0.02 0.00 -0.11 0.14 0.04 0.84 0.159
Q4_19 0.57 0.01 0.22 -0.04 -0.02 -0.05 0.21 -0.02 0.13 -0.06 0.72 0.275
Q5_1 0.14 -0.17 -0.06 0.15 0.27 0.21 0.24 0.05 0.13 -0.06 0.56 0.436
Q5_2 -0.13 0.04 0.20 -0.09 0.35 0.11 0.54 -0.10 0.10 0.20 0.66 0.342
Q5_3 -0.11 -0.05 -0.09 0.01 0.44 0.19 0.60 -0.05 0.14 0.05 0.75 0.253
Q5_4 0.04 0.01 0.12 -0.11 0.85 -0.12 0.05 0.02 0.01 0.06 0.73 0.272
Q5_5 0.11 0.03 0.06 -0.23 0.97 -0.15 -0.07 0.06 -0.02 -0.09 0.76 0.239
Q5_6 0.07 0.00 0.02 0.09 0.61 -0.01 0.30 -0.05 -0.11 0.00 0.73 0.274
Q5_7 -0.04 0.16 0.01 0.65 0.31 -0.23 -0.05 -0.04 -0.01 -0.06 0.60 0.399
Q5_8 -0.09 -0.01 0.04 0.27 0.61 0.00 0.15 -0.05 -0.02 0.00 0.67 0.333
Q5_9 0.07 -0.01 -0.22 0.51 0.37 0.14 0.11 -0.05 -0.03 0.05 0.72 0.281
Q5_10 0.09 -0.05 0.02 0.42 0.30 0.00 0.06 -0.16 0.01 -0.07 0.52 0.479
Q5_11 0.15 0.05 -0.07 0.13 0.21 0.26 0.09 0.23 0.02 -0.02 0.56 0.437
Q5_12 -0.06 0.08 0.19 0.02 0.27 0.20 0.24 -0.01 -0.02 0.05 0.52 0.478
Q6_1 -0.03 0.80 -0.24 -0.02 -0.03 0.15 0.14 0.18 0.10 0.03 0.78 0.222
Q6_2 -0.06 0.97 -0.02 -0.14 0.05 0.06 0.06 0.06 0.02 0.12 0.85 0.154
Q6_3 0.00 0.92 0.00 0.02 0.14 -0.10 -0.11 0.00 0.07 0.06 0.82 0.181
Q6_4 -0.04 0.94 0.07 -0.16 0.01 -0.04 0.11 -0.07 -0.06 0.03 0.74 0.256
Q6_5 0.01 0.19 -0.15 0.34 0.08 0.19 -0.20 0.21 0.16 -0.10 0.43 0.575
Q6_6 0.00 0.52 0.03 0.34 -0.05 0.18 -0.14 -0.14 0.09 -0.03 0.79 0.214
Q6_7 0.11 0.55 0.02 0.43 -0.10 -0.03 -0.09 -0.05 0.04 -0.04 0.80 0.197
Q6_8 -0.06 0.60 0.04 0.40 -0.15 0.08 -0.03 -0.08 -0.02 -0.05 0.78 0.216
Q6_9 -0.05 -0.08 0.09 0.79 -0.21 -0.08 -0.01 0.30 0.12 -0.01 0.57 0.431
Q6_10 -0.03 -0.05 -0.05 0.74 -0.10 0.15 0.03 0.18 -0.12 0.14 0.52 0.478
Q6_11 -0.05 0.06 0.19 0.69 -0.04 0.06 -0.01 0.04 0.01 0.28 0.70 0.298
Q7_1 -0.02 0.12 0.07 0.07 0.11 0.58 0.04 0.06 0.08 0.02 0.75 0.251
Q7_2 0.02 0.06 0.41 0.11 0.15 0.21 -0.11 -0.02 0.09 -0.04 0.61 0.389
Q7_3 -0.10 0.04 0.89 0.00 0.11 -0.14 -0.07 0.13 0.06 0.16 0.72 0.282
Q7_4 -0.10 0.07 0.68 -0.09 -0.07 0.46 -0.04 -0.15 -0.10 0.08 0.68 0.324
Q7_5 0.05 -0.04 0.57 0.12 -0.12 0.29 0.03 0.05 -0.04 -0.01 0.71 0.291
Q7_6 0.03 0.12 0.58 0.01 -0.02 0.02 0.09 0.13 0.07 -0.05 0.62 0.376
Q7_7 0.02 -0.14 0.84 -0.03 0.10 -0.13 0.07 0.24 0.07 0.12 0.79 0.205
Q7_8 0.02 -0.14 0.54 0.17 0.00 0.18 -0.02 0.02 -0.03 -0.18 0.63 0.367
Q7_9 0.09 0.02 0.05 -0.08 -0.05 0.76 0.15 0.04 -0.05 0.01 0.73 0.274
Q7_10 0.19 0.07 0.07 -0.13 -0.07 0.79 0.04 -0.08 0.00 0.03 0.78 0.216
Q7_11 0.24 0.02 0.28 0.00 0.07 0.38 -0.14 0.11 -0.19 -0.30 0.77 0.229
Q7_12 0.01 0.03 0.22 0.13 0.09 0.07 -0.07 0.59 0.03 -0.15 0.67 0.327
Q7_13 -0.07 0.00 0.22 0.22 -0.08 -0.06 -0.07 0.69 -0.05 0.07 0.58 0.422
Q7_14 -0.04 -0.06 0.55 0.05 0.13 -0.07 -0.01 0.33 -0.10 0.18 0.52 0.478
Q7_15 0.12 -0.03 0.01 0.10 -0.10 0.75 0.03 -0.02 -0.06 0.14 0.67 0.330
com
Q4_1 1.3
Q4_2 1.3
Q4_3 1.2
Q4_4 1.3
Q4_5 2.3
Q4_6 2.6
Q4_7 3.7
Q4_8 1.1
Q4_9 2.7
Q4_10 1.4
Q4_11 1.3
Q4_12 3.8
Q4_13 1.7
Q4_14 3.7
Q4_15 1.9
Q4_16 2.7
Q4_17 1.4
Q4_18 1.2
Q4_19 1.8
Q5_1 5.9
Q5_2 3.0
Q5_3 2.4
Q5_4 1.1
Q5_5 1.2
Q5_6 1.6
Q5_7 1.9
Q5_8 1.6
Q5_9 2.7
Q5_10 2.4
Q5_11 4.7
Q5_12 4.1
Q6_1 1.5
Q6_2 1.1
Q6_3 1.1
Q6_4 1.1
Q6_5 5.5
Q6_6 2.5
Q6_7 2.2
Q6_8 2.1
Q6_9 1.6
Q6_10 1.4
Q6_11 1.5
Q7_1 1.3
Q7_2 2.4
Q7_3 1.2
Q7_4 2.1
Q7_5 1.8
Q7_6 1.3
Q7_7 1.4
Q7_8 1.9
Q7_9 1.2
Q7_10 1.3
Q7_11 5.0
Q7_12 1.7
Q7_13 1.6
Q7_14 2.2
Q7_15 1.2
PA3 PA2 PA6 PA10 PA4 PA5 PA9 PA7 PA1 PA8
SS loadings 9.78 5.08 5.07 4.95 4.15 4.30 2.11 1.69 1.50 0.71
Proportion Var 0.17 0.09 0.09 0.09 0.07 0.08 0.04 0.03 0.03 0.01
Cumulative Var 0.17 0.26 0.35 0.44 0.51 0.58 0.62 0.65 0.68 0.69
Proportion Explained 0.25 0.13 0.13 0.13 0.11 0.11 0.05 0.04 0.04 0.02
Cumulative Proportion 0.25 0.38 0.51 0.63 0.74 0.85 0.90 0.94 0.98 1.00
With factor correlations of
PA3 PA2 PA6 PA10 PA4 PA5 PA9 PA7 PA1 PA8
PA3 1.00 0.54 0.62 0.71 0.51 0.67 0.58 0.14 0.27 -0.21
PA2 0.54 1.00 0.43 0.67 0.36 0.57 0.44 -0.02 0.31 -0.16
PA6 0.62 0.43 1.00 0.68 0.55 0.59 0.42 0.24 0.14 -0.18
PA10 0.71 0.67 0.68 1.00 0.55 0.73 0.57 0.10 0.34 -0.22
PA4 0.51 0.36 0.55 0.55 1.00 0.58 0.36 0.19 0.15 0.07
PA5 0.67 0.57 0.59 0.73 0.58 1.00 0.44 0.18 0.30 -0.16
PA9 0.58 0.44 0.42 0.57 0.36 0.44 1.00 0.16 0.21 -0.25
PA7 0.14 -0.02 0.24 0.10 0.19 0.18 0.16 1.00 0.02 0.03
PA1 0.27 0.31 0.14 0.34 0.15 0.30 0.21 0.02 1.00 0.07
PA8 -0.21 -0.16 -0.18 -0.22 0.07 -0.16 -0.25 0.03 0.07 1.00
Mean item complexity = 2.1
Test of the hypothesis that 10 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 84.54
The degrees of freedom for the model are 1071 and the objective function was 35.1
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.03
Fit based upon off diagonal values = 1print(fit$loadings, cutoff=0.3)
Loadings:
PA3 PA2 PA6 PA10 PA4 PA5 PA9 PA7 PA1 PA8
Q4_1 0.961
Q4_2 0.840
Q4_3 0.987
Q4_4 0.892
Q4_5 0.502
Q4_6 0.326 0.522
Q4_7 0.361 0.420
Q4_8 0.855
Q4_9 0.619 -0.340
Q4_10 0.844
Q4_11 0.640
Q4_12 0.470
Q4_13 0.334 0.688
Q4_14 0.373 0.365
Q4_15 0.627
Q4_16 0.619 0.313
Q4_17 0.630
Q4_18 0.863
Q4_19 0.569
Q5_1
Q5_2 0.349 0.537
Q5_3 0.441 0.596
Q5_4 0.850
Q5_5 0.973
Q5_6 0.610 0.301
Q5_7 0.655 0.309
Q5_8 0.612
Q5_9 0.513 0.372
Q5_10 0.419
Q5_11
Q5_12
Q6_1 0.805
Q6_2 0.970
Q6_3 0.918
Q6_4 0.937
Q6_5 0.338
Q6_6 0.518 0.336
Q6_7 0.550 0.428
Q6_8 0.595 0.402
Q6_9 0.791
Q6_10 0.742
Q6_11 0.692
Q7_1 0.581
Q7_2 0.412
Q7_3 0.885
Q7_4 0.682 0.464
Q7_5 0.573
Q7_6 0.580
Q7_7 0.835
Q7_8 0.544
Q7_9 0.763
Q7_10 0.795
Q7_11 0.375 -0.302
Q7_12 0.590
Q7_13 0.691
Q7_14 0.549 0.326
Q7_15 0.746
PA3 PA2 PA6 PA10 PA4 PA5 PA9 PA7 PA1 PA8
SS loadings 8.886 4.587 4.346 4.158 3.566 3.442 1.599 1.515 1.205 1.076
Proportion Var 0.156 0.080 0.076 0.073 0.063 0.060 0.028 0.027 0.021 0.019
Cumulative Var 0.156 0.236 0.313 0.386 0.448 0.509 0.537 0.563 0.584 0.603fit <- psych::fa(r = cov.dat, nfactors = 10, rotate = "Promax", fm = "pa", covar = T)
fitFactor Analysis using method = pa
Call: psych::fa(r = cov.dat, nfactors = 10, rotate = "Promax", covar = T,
fm = "pa")
Unstandardized loadings (pattern matrix) based upon covariance matrix
PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8 h2 u2
Q4_1 0.73 -0.14 -0.14 -0.04 0.20 0.05 -0.10 0.05 -0.05 0.15 0.49 0.26
Q4_2 0.64 -0.10 -0.03 -0.11 0.18 0.08 -0.04 -0.02 0.16 0.11 0.46 0.22
Q4_3 0.74 -0.18 -0.08 -0.01 0.12 0.09 -0.12 0.08 -0.06 0.05 0.45 0.25
Q4_4 0.61 -0.07 0.12 -0.05 0.11 0.10 -0.23 -0.06 -0.03 0.11 0.46 0.21
Q4_5 0.42 0.00 -0.02 -0.04 0.24 0.04 0.07 -0.07 0.20 0.15 0.45 0.33
Q4_6 0.25 -0.03 0.14 0.12 0.16 -0.06 0.11 0.05 -0.06 0.47 0.59 0.37
Q4_7 0.14 0.09 0.36 0.07 -0.12 0.00 0.00 -0.02 -0.10 0.34 0.41 0.47
Q4_8 0.79 -0.03 -0.04 0.02 -0.01 0.00 -0.04 -0.05 0.14 0.02 0.53 0.33
Q4_9 0.77 -0.27 0.21 0.06 -0.04 -0.04 0.14 0.02 0.16 -0.19 0.59 0.42
Q4_10 0.58 0.06 0.12 0.01 -0.07 0.08 -0.24 -0.02 -0.03 0.10 0.37 0.25
Q4_11 0.65 0.21 0.03 -0.02 -0.04 -0.05 -0.03 0.04 0.05 0.03 0.56 0.35
Q4_12 0.39 0.30 0.19 0.13 -0.16 -0.03 0.13 -0.08 0.52 0.01 0.74 0.37
Q4_13 0.37 0.07 -0.06 0.10 0.03 0.09 0.17 -0.01 0.52 -0.10 0.53 0.23
Q4_14 0.44 0.36 -0.05 0.04 -0.11 -0.10 0.12 0.13 0.04 0.05 0.45 0.35
Q4_15 0.69 0.26 -0.14 0.00 -0.12 -0.04 0.07 0.02 -0.04 -0.06 0.57 0.27
Q4_16 0.69 0.08 -0.21 0.17 -0.15 -0.05 0.12 0.01 -0.21 0.08 0.57 0.32
Q4_17 0.69 0.05 0.05 0.07 -0.05 -0.13 0.00 -0.05 0.04 -0.01 0.48 0.45
Q4_18 0.70 0.07 -0.04 -0.12 0.04 0.05 -0.04 -0.09 0.12 0.06 0.50 0.17
Q4_19 0.70 -0.04 0.18 0.00 -0.05 -0.08 0.18 -0.02 0.12 -0.08 0.63 0.31
Q5_1 0.19 0.16 -0.06 -0.16 0.23 0.06 0.35 0.05 0.06 -0.03 0.46 0.44
Q5_2 -0.02 -0.09 0.21 0.05 0.07 0.10 0.61 -0.07 0.08 0.16 0.66 0.41
Q5_3 0.03 0.02 -0.12 -0.05 0.18 0.14 0.77 -0.01 0.13 0.03 0.78 0.31
Q5_4 -0.02 -0.13 0.13 0.02 -0.10 0.94 0.10 0.00 0.05 0.04 0.87 0.36
Q5_5 0.02 -0.21 0.05 0.02 -0.12 1.10 -0.04 0.03 0.00 -0.12 0.85 0.22
Q5_6 0.09 0.10 0.01 0.00 -0.02 0.37 0.27 -0.05 -0.10 -0.02 0.55 0.27
Q5_7 -0.01 0.66 0.00 0.11 -0.21 0.20 -0.05 -0.07 -0.02 -0.05 0.49 0.39
Q5_8 -0.08 0.28 0.04 -0.02 0.04 0.42 0.25 -0.07 -0.04 0.02 0.68 0.44
Q5_9 0.06 0.49 -0.20 -0.02 0.18 0.14 0.18 -0.05 -0.07 0.08 0.65 0.34
Q5_10 0.10 0.31 0.03 -0.04 0.00 0.11 0.08 -0.14 -0.03 -0.01 0.31 0.39
Q5_11 0.16 0.12 -0.07 0.04 0.30 0.07 0.14 0.21 -0.04 0.00 0.46 0.45
Q5_12 0.02 0.00 0.18 0.08 0.18 0.08 0.26 0.01 -0.05 0.05 0.44 0.47
Q6_1 0.01 -0.03 -0.17 0.66 0.09 -0.03 0.11 0.13 0.06 0.04 0.45 0.26
Q6_2 -0.03 -0.19 -0.01 0.90 0.06 0.00 0.04 0.07 0.00 0.11 0.65 0.18
Q6_3 -0.05 -0.03 -0.02 0.83 -0.05 0.14 -0.09 0.02 0.07 0.07 0.61 0.25
Q6_4 0.03 -0.23 0.06 0.87 -0.04 -0.02 0.04 -0.05 -0.05 0.01 0.58 0.28
Q6_5 -0.06 0.42 -0.15 0.13 0.25 0.17 -0.14 0.19 0.15 -0.08 0.39 0.68
Q6_6 -0.01 0.20 0.00 0.43 0.15 0.05 -0.13 -0.09 0.13 -0.04 0.45 0.22
Q6_7 0.11 0.33 0.01 0.47 -0.01 -0.05 -0.13 -0.05 0.06 -0.04 0.54 0.23
Q6_8 -0.01 0.32 0.01 0.52 0.08 -0.11 -0.10 -0.07 0.04 -0.06 0.57 0.25
Q6_9 0.03 1.05 0.10 -0.12 -0.11 -0.25 -0.02 0.33 0.16 -0.03 0.74 0.75
Q6_10 -0.03 0.64 -0.03 -0.04 0.12 -0.06 -0.06 0.14 -0.04 0.08 0.38 0.46
Q6_11 -0.09 0.64 0.22 0.04 0.03 -0.07 -0.01 0.01 0.07 0.24 0.61 0.33
Q7_1 -0.04 0.05 0.06 0.08 0.55 0.04 0.12 0.04 0.03 0.03 0.54 0.27
Q7_2 -0.01 0.12 0.33 0.01 0.21 0.15 -0.05 -0.05 0.06 -0.05 0.41 0.33
Q7_3 -0.09 0.03 0.86 0.03 -0.12 0.10 -0.04 0.10 0.06 0.14 0.68 0.34
Q7_4 -0.10 -0.15 0.63 0.06 0.44 -0.12 -0.04 -0.16 -0.08 0.07 0.61 0.35
Q7_5 0.08 0.11 0.52 -0.05 0.28 -0.13 -0.02 0.01 -0.04 -0.02 0.60 0.30
Q7_6 0.10 0.11 0.51 0.08 0.01 -0.05 0.05 0.08 0.05 -0.05 0.49 0.36
Q7_7 0.06 -0.02 0.82 -0.09 -0.12 0.13 0.03 0.18 0.10 0.06 0.72 0.31
Q7_8 0.08 0.18 0.45 -0.11 0.14 -0.01 -0.06 -0.02 -0.02 -0.15 0.44 0.33
Q7_9 0.09 -0.12 0.03 0.00 0.78 -0.10 0.14 0.03 -0.06 0.01 0.65 0.34
Q7_10 0.14 -0.16 0.01 0.07 0.77 -0.06 0.03 -0.06 0.02 0.01 0.62 0.27
Q7_11 0.20 -0.06 0.21 0.02 0.33 0.10 -0.15 0.07 -0.19 -0.23 0.48 0.22
Q7_12 0.03 0.17 0.22 0.04 0.14 0.14 -0.04 0.49 -0.05 -0.11 0.60 0.45
Q7_13 -0.07 0.30 0.19 0.02 -0.06 -0.04 -0.03 0.66 -0.06 0.08 0.53 0.45
Q7_14 -0.06 0.04 0.54 -0.01 -0.08 0.10 0.01 0.28 -0.07 0.19 0.47 0.50
Q7_15 0.06 0.01 0.00 -0.03 0.68 -0.11 0.05 -0.01 -0.04 0.11 0.48 0.34
H2 U2
Q4_1 0.65 0.35
Q4_2 0.68 0.32
Q4_3 0.64 0.36
Q4_4 0.69 0.31
Q4_5 0.58 0.42
Q4_6 0.61 0.39
Q4_7 0.46 0.54
Q4_8 0.61 0.39
Q4_9 0.59 0.41
Q4_10 0.60 0.40
Q4_11 0.61 0.39
Q4_12 0.67 0.33
Q4_13 0.70 0.30
Q4_14 0.56 0.44
Q4_15 0.68 0.32
Q4_16 0.64 0.36
Q4_17 0.52 0.48
Q4_18 0.74 0.26
Q4_19 0.67 0.33
Q5_1 0.51 0.49
Q5_2 0.62 0.38
Q5_3 0.72 0.28
Q5_4 0.71 0.29
Q5_5 0.79 0.21
Q5_6 0.67 0.33
Q5_7 0.56 0.44
Q5_8 0.61 0.39
Q5_9 0.65 0.35
Q5_10 0.45 0.55
Q5_11 0.51 0.49
Q5_12 0.49 0.51
Q6_1 0.63 0.37
Q6_2 0.78 0.22
Q6_3 0.71 0.29
Q6_4 0.67 0.33
Q6_5 0.36 0.64
Q6_6 0.67 0.33
Q6_7 0.70 0.30
Q6_8 0.70 0.30
Q6_9 0.50 0.50
Q6_10 0.45 0.55
Q6_11 0.65 0.35
Q7_1 0.67 0.33
Q7_2 0.56 0.44
Q7_3 0.66 0.34
Q7_4 0.63 0.37
Q7_5 0.67 0.33
Q7_6 0.58 0.42
Q7_7 0.70 0.30
Q7_8 0.58 0.42
Q7_9 0.66 0.34
Q7_10 0.70 0.30
Q7_11 0.69 0.31
Q7_12 0.57 0.43
Q7_13 0.54 0.46
Q7_14 0.49 0.51
Q7_15 0.59 0.41
PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8
SS loadings 7.80 4.05 4.28 3.72 3.71 2.91 2.21 1.13 0.63 0.85
Proportion Var 0.15 0.08 0.08 0.07 0.07 0.06 0.04 0.02 0.01 0.02
Cumulative Var 0.15 0.23 0.32 0.39 0.46 0.52 0.57 0.59 0.60 0.62
Proportion Explained 0.25 0.13 0.14 0.12 0.12 0.09 0.07 0.04 0.02 0.03
Cumulative Proportion 0.25 0.38 0.52 0.63 0.75 0.85 0.92 0.95 0.97 1.00
Standardized loadings (pattern matrix)
item PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8 h2
Q4_1 1 0.84 -0.16 -0.17 -0.05 0.23 0.05 -0.12 0.06 -0.05 0.17 0.65
Q4_2 2 0.78 -0.12 -0.04 -0.13 0.22 0.10 -0.04 -0.02 0.19 0.13 0.68
Q4_3 3 0.88 -0.21 -0.10 -0.02 0.15 0.11 -0.14 0.09 -0.07 0.05 0.64
Q4_4 4 0.75 -0.09 0.15 -0.06 0.13 0.12 -0.28 -0.08 -0.04 0.14 0.69
Q4_5 5 0.48 0.00 -0.02 -0.05 0.27 0.05 0.08 -0.08 0.22 0.17 0.58
Q4_6 6 0.25 -0.03 0.14 0.13 0.16 -0.06 0.11 0.05 -0.06 0.48 0.61
Q4_7 7 0.15 0.10 0.39 0.08 -0.13 0.00 0.00 -0.02 -0.11 0.37 0.46
Q4_8 8 0.85 -0.03 -0.05 0.02 -0.01 0.00 -0.04 -0.05 0.15 0.02 0.61
Q4_9 9 0.76 -0.27 0.21 0.06 -0.04 -0.04 0.14 0.02 0.16 -0.19 0.59
Q4_10 10 0.73 0.08 0.15 0.01 -0.09 0.10 -0.31 -0.03 -0.04 0.12 0.60
Q4_11 11 0.68 0.22 0.03 -0.02 -0.04 -0.05 -0.03 0.04 0.05 0.04 0.61
Q4_12 12 0.37 0.28 0.19 0.12 -0.15 -0.03 0.12 -0.08 0.49 0.01 0.67
Q4_13 13 0.42 0.08 -0.06 0.11 0.03 0.11 0.19 -0.01 0.60 -0.12 0.70
Q4_14 14 0.49 0.40 -0.05 0.04 -0.12 -0.11 0.13 0.15 0.04 0.06 0.56
Q4_15 15 0.76 0.28 -0.16 0.00 -0.14 -0.04 0.08 0.02 -0.04 -0.06 0.68
Q4_16 16 0.73 0.09 -0.22 0.18 -0.16 -0.06 0.13 0.01 -0.22 0.08 0.64
Q4_17 17 0.71 0.05 0.05 0.07 -0.05 -0.14 0.00 -0.06 0.05 -0.01 0.52
Q4_18 18 0.85 0.09 -0.04 -0.14 0.05 0.06 -0.05 -0.10 0.15 0.07 0.74
Q4_19 19 0.72 -0.04 0.19 0.00 -0.05 -0.08 0.18 -0.03 0.13 -0.08 0.67
Q5_1 20 0.20 0.17 -0.06 -0.17 0.24 0.06 0.37 0.05 0.06 -0.04 0.51
Q5_2 21 -0.02 -0.09 0.21 0.05 0.07 0.10 0.59 -0.07 0.08 0.16 0.62
Q5_3 22 0.03 0.02 -0.11 -0.05 0.17 0.13 0.73 -0.01 0.12 0.03 0.72
Q5_4 23 -0.02 -0.12 0.12 0.02 -0.09 0.85 0.09 0.00 0.05 0.04 0.71
Q5_5 24 0.02 -0.20 0.05 0.02 -0.12 1.06 -0.04 0.02 0.00 -0.11 0.79
Q5_6 25 0.10 0.11 0.01 0.00 -0.02 0.41 0.29 -0.06 -0.11 -0.02 0.67
Q5_7 26 -0.01 0.70 0.00 0.12 -0.22 0.22 -0.05 -0.07 -0.02 -0.05 0.56
Q5_8 27 -0.08 0.26 0.04 -0.02 0.03 0.40 0.23 -0.07 -0.03 0.01 0.61
Q5_9 28 0.06 0.49 -0.20 -0.02 0.18 0.14 0.18 -0.06 -0.07 0.08 0.65
Q5_10 29 0.11 0.38 0.04 -0.05 0.00 0.13 0.10 -0.16 -0.04 -0.02 0.45
Q5_11 30 0.17 0.12 -0.08 0.05 0.32 0.07 0.15 0.22 -0.05 0.00 0.51
Q5_12 31 0.02 0.00 0.19 0.09 0.19 0.08 0.28 0.01 -0.05 0.05 0.49
Q6_1 32 0.01 -0.04 -0.20 0.78 0.11 -0.04 0.13 0.16 0.07 0.05 0.63
Q6_2 33 -0.04 -0.20 -0.01 0.98 0.06 0.00 0.05 0.07 0.01 0.12 0.78
Q6_3 34 -0.05 -0.03 -0.02 0.90 -0.06 0.15 -0.09 0.02 0.07 0.08 0.71
Q6_4 35 0.03 -0.25 0.06 0.94 -0.05 -0.03 0.04 -0.05 -0.05 0.01 0.67
Q6_5 36 -0.06 0.40 -0.14 0.12 0.24 0.16 -0.14 0.18 0.15 -0.07 0.36
Q6_6 37 -0.01 0.24 0.01 0.53 0.18 0.06 -0.15 -0.11 0.16 -0.05 0.67
Q6_7 38 0.12 0.38 0.01 0.54 -0.01 -0.06 -0.15 -0.06 0.07 -0.05 0.70
Q6_8 39 -0.01 0.35 0.01 0.58 0.09 -0.12 -0.11 -0.08 0.05 -0.07 0.70
Q6_9 40 0.02 0.86 0.08 -0.10 -0.09 -0.20 -0.01 0.27 0.13 -0.03 0.50
Q6_10 41 -0.03 0.70 -0.03 -0.04 0.13 -0.07 -0.07 0.15 -0.04 0.08 0.45
Q6_11 42 -0.09 0.66 0.23 0.04 0.03 -0.07 -0.01 0.01 0.07 0.24 0.65
Q7_1 43 -0.05 0.06 0.06 0.09 0.61 0.04 0.13 0.04 0.03 0.03 0.67
Q7_2 44 -0.01 0.14 0.39 0.02 0.24 0.17 -0.06 -0.05 0.07 -0.05 0.56
Q7_3 45 -0.09 0.03 0.85 0.03 -0.12 0.10 -0.04 0.09 0.06 0.14 0.66
Q7_4 46 -0.10 -0.15 0.65 0.07 0.45 -0.12 -0.04 -0.16 -0.08 0.07 0.63
Q7_5 47 0.09 0.12 0.55 -0.05 0.30 -0.14 -0.02 0.02 -0.04 -0.02 0.67
Q7_6 48 0.11 0.12 0.55 0.09 0.01 -0.06 0.05 0.09 0.05 -0.06 0.58
Q7_7 49 0.06 -0.02 0.81 -0.09 -0.12 0.13 0.03 0.17 0.10 0.06 0.70
Q7_8 50 0.09 0.20 0.51 -0.13 0.16 -0.01 -0.07 -0.02 -0.02 -0.17 0.58
Q7_9 51 0.09 -0.12 0.03 0.00 0.79 -0.10 0.14 0.03 -0.06 0.01 0.66
Q7_10 52 0.15 -0.17 0.01 0.08 0.82 -0.07 0.03 -0.06 0.02 0.01 0.70
Q7_11 53 0.24 -0.07 0.25 0.03 0.40 0.11 -0.18 0.08 -0.22 -0.27 0.69
Q7_12 54 0.02 0.17 0.22 0.04 0.14 0.13 -0.04 0.48 -0.05 -0.11 0.57
Q7_13 55 -0.07 0.30 0.19 0.02 -0.06 -0.04 -0.04 0.66 -0.06 0.09 0.54
Q7_14 56 -0.06 0.04 0.55 -0.02 -0.09 0.10 0.01 0.28 -0.07 0.19 0.49
Q7_15 57 0.06 0.01 0.00 -0.03 0.75 -0.12 0.05 -0.01 -0.05 0.12 0.59
u2
Q4_1 0.35
Q4_2 0.32
Q4_3 0.36
Q4_4 0.31
Q4_5 0.42
Q4_6 0.39
Q4_7 0.54
Q4_8 0.39
Q4_9 0.41
Q4_10 0.40
Q4_11 0.39
Q4_12 0.33
Q4_13 0.30
Q4_14 0.44
Q4_15 0.32
Q4_16 0.36
Q4_17 0.48
Q4_18 0.26
Q4_19 0.33
Q5_1 0.49
Q5_2 0.38
Q5_3 0.28
Q5_4 0.29
Q5_5 0.21
Q5_6 0.33
Q5_7 0.44
Q5_8 0.39
Q5_9 0.35
Q5_10 0.55
Q5_11 0.49
Q5_12 0.51
Q6_1 0.37
Q6_2 0.22
Q6_3 0.29
Q6_4 0.33
Q6_5 0.64
Q6_6 0.33
Q6_7 0.30
Q6_8 0.30
Q6_9 0.50
Q6_10 0.55
Q6_11 0.35
Q7_1 0.33
Q7_2 0.44
Q7_3 0.34
Q7_4 0.37
Q7_5 0.33
Q7_6 0.42
Q7_7 0.30
Q7_8 0.42
Q7_9 0.34
Q7_10 0.30
Q7_11 0.31
Q7_12 0.43
Q7_13 0.46
Q7_14 0.51
Q7_15 0.41
PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8
SS loadings 8.76 4.55 4.81 4.18 4.17 3.26 2.48 1.27 0.71 0.95
Proportion Var 0.15 0.08 0.08 0.07 0.07 0.06 0.04 0.02 0.01 0.02
Cumulative Var 0.15 0.23 0.32 0.39 0.46 0.52 0.57 0.59 0.60 0.62
Cum. factor Var 0.25 0.38 0.52 0.63 0.75 0.85 0.92 0.95 0.97 1.00
With factor correlations of
PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8
PA1 1.00 0.73 0.59 0.55 0.71 0.56 0.56 -0.05 -0.17 0.08
PA9 0.73 1.00 0.67 0.69 0.76 0.60 0.62 -0.12 -0.21 0.06
PA2 0.59 0.67 1.00 0.42 0.60 0.57 0.49 0.07 -0.32 0.02
PA3 0.55 0.69 0.42 1.00 0.60 0.39 0.45 -0.18 -0.05 0.02
PA6 0.71 0.76 0.60 0.60 1.00 0.57 0.48 -0.03 -0.15 0.07
PA4 0.56 0.60 0.57 0.39 0.57 1.00 0.59 -0.04 -0.37 0.26
PA5 0.56 0.62 0.49 0.45 0.48 0.59 1.00 -0.07 -0.31 0.06
PA7 -0.05 -0.12 0.07 -0.18 -0.03 -0.04 -0.07 1.00 0.07 -0.02
PA10 -0.17 -0.21 -0.32 -0.05 -0.15 -0.37 -0.31 0.07 1.00 0.03
PA8 0.08 0.06 0.02 0.02 0.07 0.26 0.06 -0.02 0.03 1.00
Mean item complexity = 1.9
Test of the hypothesis that 10 factors are sufficient.
The degrees of freedom for the null model are 1596 and the objective function was 46.33
The degrees of freedom for the model are 1071 and the objective function was 6.24
The root mean square of the residuals (RMSR) is 0.02
The df corrected root mean square of the residuals is 0.02
Fit based upon off diagonal values = 1
Measures of factor score adequacy
PA1 PA9 PA2 PA3 PA6 PA4
Correlation of (regression) scores with factors 0.98 0.96 0.96 0.97 0.96 0.96
Multiple R square of scores with factors 0.96 0.92 0.92 0.94 0.92 0.92
Minimum correlation of possible factor scores 0.92 0.85 0.84 0.88 0.84 0.83
PA5 PA7 PA10 PA8
Correlation of (regression) scores with factors 0.92 0.85 0.86 0.83
Multiple R square of scores with factors 0.85 0.72 0.74 0.69
Minimum correlation of possible factor scores 0.71 0.45 0.48 0.38print(fit$loadings, cutoff=0.3)
Loadings:
PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8
Q4_1 0.730
Q4_2 0.643
Q4_3 0.737
Q4_4 0.609
Q4_5 0.420
Q4_6 0.470
Q4_7 0.363 0.344
Q4_8 0.788
Q4_9 0.767
Q4_10 0.580
Q4_11 0.654
Q4_12 0.387 0.519
Q4_13 0.370 0.525
Q4_14 0.435 0.359
Q4_15 0.694
Q4_16 0.686
Q4_17 0.689
Q4_18 0.699
Q4_19 0.697
Q5_1 0.349
Q5_2 0.608
Q5_3 0.767
Q5_4 0.936
Q5_5 1.095
Q5_6 0.370
Q5_7 0.657
Q5_8 0.419
Q5_9 0.485
Q5_10 0.315
Q5_11 0.304
Q5_12
Q6_1 0.660
Q6_2 0.895
Q6_3 0.832
Q6_4 0.869
Q6_5 0.418
Q6_6 0.434
Q6_7 0.333 0.471
Q6_8 0.319 0.522
Q6_9 1.051 0.326
Q6_10 0.643
Q6_11 0.641
Q7_1 0.551
Q7_2 0.331
Q7_3 0.861
Q7_4 0.633 0.444
Q7_5 0.519
Q7_6 0.510
Q7_7 0.817
Q7_8 0.446
Q7_9 0.783
Q7_10 0.771
Q7_11 0.335
Q7_12 0.494
Q7_13 0.657
Q7_14 0.541
Q7_15 0.675
PA1 PA9 PA2 PA3 PA6 PA4 PA5 PA7 PA10 PA8
SS loadings 7.214 4.173 3.751 3.625 3.218 2.886 1.809 1.188 0.971 0.809
Proportion Var 0.127 0.073 0.066 0.064 0.056 0.051 0.032 0.021 0.017 0.014
Cumulative Var 0.127 0.200 0.266 0.329 0.386 0.436 0.468 0.489 0.506 0.520Selected Set of Items
We selected the following items based on these results:
| Effective Learning (Q4) | Student Centered (Q5) | Interactive (Q6) | Engagement (Q7) |
|---|---|---|---|
| Q4-1 | Q5-1 | Q6-1 | Q7-2 |
| Q4-2 | Q5-2 | Q6-2 | Q7-4 |
| Q4-3 | Q5-3 | Q6-3 | Q7-5 |
| Q4-4 | Q5-4 | Q6-4 | Q7-7 |
| Q4-5 | Q5-5 | Q6-5 | Q7-8 |
| Q4-8 | Q5-6 | Q6-6 | Q7-12 |
| Q4-9 | Q5-8 | Q6-7 | Q7-13 |
| Q4-10 | Q5-12 | Q6-8 | Q7-14 |
| Q4-11 | Q6-12 | ||
| Q4-15 | |||
| Q4-16 | |||
| Q4-17 | |||
| Q4-18 | |||
| Q4-19 |
For the item stems, please see the document here.
Post EFA Item & Reliability Analysis
Effective Learning Items
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q4_",c(1:5,8:11, 15:19))])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q4_1 | 0.866 | 0.767 | 0.726 | 2.460 | 0.663 | 0.628 |
| Q4_2 | 0.819 | 0.801 | 0.767 | 2.305 | 0.655 | 0.628 |
| Q4_3 | 0.834 | 0.781 | 0.744 | 2.537 | 0.651 | 0.620 |
| Q4_4 | 0.816 | 0.786 | 0.750 | 2.640 | 0.640 | 0.611 |
| Q4_5 | 0.880 | 0.723 | 0.675 | 2.320 | 0.636 | 0.594 |
| Q4_8 | 0.927 | 0.794 | 0.754 | 2.287 | 0.735 | 0.698 |
| Q4_9 | 1.007 | 0.719 | 0.662 | 2.463 | 0.723 | 0.666 |
| Q4_10 | 0.789 | 0.745 | 0.705 | 2.582 | 0.587 | 0.556 |
| Q4_11 | 0.955 | 0.792 | 0.750 | 2.500 | 0.755 | 0.715 |
| Q4_15 | 0.917 | 0.793 | 0.753 | 2.375 | 0.726 | 0.690 |
| Q4_16 | 0.941 | 0.732 | 0.681 | 2.457 | 0.687 | 0.640 |
| Q4_17 | 0.964 | 0.734 | 0.682 | 2.198 | 0.707 | 0.657 |
| Q4_18 | 0.823 | 0.851 | 0.825 | 2.360 | 0.700 | 0.678 |
| Q4_19 | 0.970 | 0.800 | 0.759 | 2.543 | 0.774 | 0.735 |
# post reliability
psych::alpha(dat[, paste0("Q4_",c(1:5,8:11, 15:19))])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q4_", c(1:5, 8:11, 15:19))])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.95 0.95 0.95 0.57 18 0.0043 2.4 0.69 0.56
lower alpha upper 95% confidence boundaries
0.94 0.95 0.96
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q4_1 0.94 0.94 0.95 0.57 17 0.0046 0.0044 0.56
Q4_2 0.94 0.94 0.95 0.56 17 0.0047 0.0045 0.56
Q4_3 0.94 0.94 0.95 0.56 17 0.0046 0.0046 0.56
Q4_4 0.94 0.94 0.95 0.56 17 0.0047 0.0045 0.56
Q4_5 0.94 0.95 0.95 0.57 17 0.0045 0.0045 0.57
Q4_8 0.94 0.94 0.95 0.56 17 0.0047 0.0050 0.55
Q4_9 0.95 0.95 0.95 0.58 18 0.0045 0.0039 0.57
Q4_10 0.94 0.94 0.95 0.57 17 0.0046 0.0048 0.56
Q4_11 0.94 0.94 0.95 0.56 17 0.0047 0.0050 0.55
Q4_15 0.94 0.94 0.95 0.56 17 0.0047 0.0049 0.56
Q4_16 0.94 0.95 0.95 0.57 17 0.0045 0.0043 0.57
Q4_17 0.94 0.95 0.95 0.57 17 0.0045 0.0047 0.57
Q4_18 0.94 0.94 0.95 0.56 16 0.0048 0.0041 0.55
Q4_19 0.94 0.94 0.95 0.56 17 0.0047 0.0047 0.56
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q4_1 328 0.77 0.77 0.75 0.73 2.5 0.87
Q4_2 328 0.80 0.81 0.79 0.77 2.3 0.82
Q4_3 328 0.78 0.79 0.77 0.74 2.5 0.83
Q4_4 328 0.79 0.79 0.78 0.75 2.6 0.82
Q4_5 328 0.72 0.73 0.70 0.68 2.3 0.88
Q4_8 328 0.79 0.79 0.77 0.75 2.3 0.93
Q4_9 328 0.72 0.71 0.69 0.66 2.5 1.01
Q4_10 328 0.75 0.75 0.73 0.71 2.6 0.79
Q4_11 328 0.79 0.79 0.77 0.75 2.5 0.95
Q4_15 328 0.79 0.79 0.77 0.75 2.4 0.92
Q4_16 328 0.73 0.73 0.71 0.68 2.5 0.94
Q4_17 328 0.73 0.73 0.70 0.68 2.2 0.96
Q4_18 328 0.85 0.85 0.85 0.82 2.4 0.82
Q4_19 328 0.80 0.79 0.78 0.76 2.5 0.97
Non missing response frequency for each item
1 2 3 4 5 miss
Q4_1 0.13 0.38 0.39 0.09 0.01 0
Q4_2 0.15 0.47 0.32 0.05 0.01 0
Q4_3 0.10 0.37 0.44 0.08 0.02 0
Q4_4 0.09 0.28 0.54 0.08 0.02 0
Q4_5 0.16 0.47 0.28 0.08 0.01 0
Q4_8 0.20 0.43 0.28 0.07 0.02 0
Q4_9 0.15 0.42 0.27 0.12 0.04 0
Q4_10 0.09 0.30 0.54 0.04 0.02 0
Q4_11 0.16 0.34 0.38 0.10 0.02 0
Q4_15 0.16 0.41 0.33 0.07 0.02 0
Q4_16 0.17 0.33 0.40 0.08 0.02 0
Q4_17 0.26 0.39 0.25 0.08 0.02 0
Q4_18 0.13 0.47 0.34 0.06 0.01 0
Q4_19 0.14 0.35 0.39 0.08 0.04 0Initial Student Centered Items
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q5_",c(1:6, 8, 12))])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q5_1 | 0.951 | 0.686 | 0.591 | 2.637 | 0.652 | 0.561 |
| Q5_2 | 1.035 | 0.778 | 0.697 | 2.954 | 0.804 | 0.720 |
| Q5_3 | 1.046 | 0.790 | 0.711 | 2.662 | 0.825 | 0.743 |
| Q5_4 | 1.106 | 0.785 | 0.700 | 3.500 | 0.867 | 0.772 |
| Q5_5 | 1.034 | 0.736 | 0.643 | 3.530 | 0.760 | 0.664 |
| Q5_6 | 0.906 | 0.827 | 0.771 | 2.927 | 0.748 | 0.697 |
| Q5_8 | 1.055 | 0.792 | 0.713 | 2.905 | 0.835 | 0.752 |
| Q5_12 | 0.952 | 0.701 | 0.608 | 2.881 | 0.666 | 0.578 |
# post reliability
psych::alpha(dat[, paste0("Q5_",c(1:6, 8, 12))])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q5_", c(1:6, 8, 12))])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.9 0.9 0.9 0.52 8.7 0.0087 3 0.77 0.51
lower alpha upper 95% confidence boundaries
0.88 0.9 0.91
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q5_1 0.89 0.89 0.90 0.54 8.3 0.0093 0.0088 0.53
Q5_2 0.88 0.88 0.89 0.52 7.5 0.0100 0.0106 0.52
Q5_3 0.88 0.88 0.88 0.51 7.4 0.0101 0.0100 0.51
Q5_4 0.88 0.88 0.87 0.52 7.5 0.0100 0.0075 0.51
Q5_5 0.89 0.89 0.88 0.53 7.9 0.0095 0.0048 0.52
Q5_6 0.88 0.88 0.88 0.50 7.0 0.0105 0.0096 0.50
Q5_8 0.88 0.88 0.89 0.51 7.4 0.0102 0.0109 0.51
Q5_12 0.89 0.89 0.90 0.54 8.1 0.0094 0.0107 0.54
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q5_1 328 0.69 0.69 0.63 0.59 2.6 0.95
Q5_2 328 0.78 0.78 0.74 0.70 3.0 1.04
Q5_3 328 0.79 0.79 0.76 0.71 2.7 1.05
Q5_4 328 0.79 0.78 0.76 0.70 3.5 1.11
Q5_5 328 0.74 0.73 0.71 0.64 3.5 1.03
Q5_6 328 0.83 0.83 0.81 0.77 2.9 0.91
Q5_8 328 0.79 0.79 0.75 0.71 2.9 1.06
Q5_12 328 0.70 0.71 0.64 0.61 2.9 0.95
Non missing response frequency for each item
1 2 3 4 5 miss
Q5_1 0.12 0.32 0.40 0.14 0.03 0
Q5_2 0.10 0.20 0.38 0.27 0.05 0
Q5_3 0.15 0.29 0.35 0.17 0.04 0
Q5_4 0.08 0.11 0.21 0.46 0.15 0
Q5_5 0.05 0.12 0.21 0.48 0.14 0
Q5_6 0.09 0.15 0.52 0.21 0.02 0
Q5_8 0.12 0.19 0.39 0.24 0.05 0
Q5_12 0.10 0.17 0.50 0.19 0.04 0Interactive Items
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q6_",c(1:8, 11))])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q6_1 | 0.844 | 0.757 | 0.690 | 1.701 | 0.638 | 0.581 |
| Q6_2 | 0.912 | 0.828 | 0.773 | 2.079 | 0.754 | 0.703 |
| Q6_3 | 0.924 | 0.824 | 0.766 | 1.930 | 0.760 | 0.707 |
| Q6_4 | 0.927 | 0.774 | 0.703 | 2.076 | 0.716 | 0.651 |
| Q6_5 | 1.033 | 0.572 | 0.444 | 2.351 | 0.589 | 0.458 |
| Q6_6 | 0.820 | 0.818 | 0.766 | 1.832 | 0.669 | 0.627 |
| Q6_7 | 0.875 | 0.841 | 0.791 | 2.162 | 0.735 | 0.691 |
| Q6_8 | 0.903 | 0.827 | 0.772 | 2.113 | 0.746 | 0.697 |
| Q6_11 | 0.970 | 0.659 | 0.557 | 2.814 | 0.638 | 0.539 |
# post reliability
psych::alpha(dat[, paste0("Q6_",c(1:8, 11))])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q6_", c(1:8, 11))])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.91 0.91 0.92 0.54 10 0.0076 2.1 0.7 0.57
lower alpha upper 95% confidence boundaries
0.89 0.91 0.92
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q6_1 0.90 0.90 0.91 0.54 9.3 0.0086 0.019 0.57
Q6_2 0.89 0.90 0.90 0.52 8.7 0.0091 0.018 0.56
Q6_3 0.89 0.90 0.90 0.52 8.7 0.0091 0.019 0.56
Q6_4 0.90 0.90 0.90 0.53 9.2 0.0087 0.017 0.56
Q6_5 0.92 0.92 0.92 0.59 11.4 0.0069 0.010 0.59
Q6_6 0.89 0.90 0.90 0.52 8.8 0.0090 0.021 0.56
Q6_7 0.89 0.90 0.90 0.52 8.6 0.0092 0.020 0.56
Q6_8 0.89 0.90 0.90 0.52 8.7 0.0091 0.020 0.56
Q6_11 0.91 0.91 0.92 0.56 10.4 0.0077 0.019 0.59
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q6_1 328 0.76 0.76 0.73 0.69 1.7 0.84
Q6_2 328 0.83 0.83 0.82 0.77 2.1 0.91
Q6_3 328 0.82 0.83 0.81 0.77 1.9 0.92
Q6_4 328 0.77 0.78 0.75 0.70 2.1 0.93
Q6_5 328 0.57 0.56 0.47 0.44 2.4 1.03
Q6_6 328 0.82 0.82 0.80 0.77 1.8 0.82
Q6_7 328 0.84 0.84 0.83 0.79 2.2 0.88
Q6_8 328 0.83 0.83 0.81 0.77 2.1 0.90
Q6_11 328 0.66 0.65 0.59 0.56 2.8 0.97
Non missing response frequency for each item
1 2 3 4 5 miss
Q6_1 0.49 0.36 0.09 0.05 0.00 0
Q6_2 0.27 0.47 0.17 0.07 0.01 0
Q6_3 0.36 0.45 0.13 0.05 0.02 0
Q6_4 0.29 0.43 0.20 0.06 0.01 0
Q6_5 0.19 0.46 0.21 0.10 0.04 0
Q6_6 0.37 0.48 0.11 0.03 0.01 0
Q6_7 0.21 0.50 0.20 0.07 0.01 0
Q6_8 0.25 0.47 0.20 0.07 0.01 0
Q6_11 0.11 0.21 0.49 0.14 0.05 0Engagement Items
# item analysis
fit <- psychometric::item.exam(dat[, paste0("Q7_",c(2, 4:5, 7:8, 12:14))])
kable(fit[,c(1:4,7:8)], format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%")| Sample.SD | Item.total | Item.Tot.woi | Difficulty | Item.Reliab | Item.Rel.woi | |
|---|---|---|---|---|---|---|
| Q7_2 | 0.857 | 0.701 | 0.607 | 2.692 | 0.599 | 0.519 |
| Q7_4 | 0.980 | 0.710 | 0.602 | 2.841 | 0.694 | 0.589 |
| Q7_5 | 0.948 | 0.798 | 0.720 | 2.875 | 0.756 | 0.682 |
| Q7_7 | 1.014 | 0.789 | 0.701 | 3.652 | 0.799 | 0.710 |
| Q7_8 | 0.877 | 0.736 | 0.648 | 2.814 | 0.644 | 0.567 |
| Q7_12 | 1.023 | 0.717 | 0.605 | 3.390 | 0.732 | 0.618 |
| Q7_13 | 0.989 | 0.571 | 0.430 | 3.579 | 0.564 | 0.425 |
| Q7_14 | 0.986 | 0.694 | 0.581 | 3.610 | 0.684 | 0.573 |
# post reliability
psych::alpha(dat[, paste0("Q7_",c(2, 4:5, 7:8, 12:14))])
Reliability analysis
Call: psych::alpha(x = dat[, paste0("Q7_", c(2, 4:5, 7:8, 12:14))])
raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
0.86 0.86 0.87 0.44 6.3 0.012 3.2 0.69 0.45
lower alpha upper 95% confidence boundaries
0.84 0.86 0.88
Reliability if an item is dropped:
raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
Q7_2 0.85 0.85 0.85 0.44 5.5 0.013 0.0148 0.45
Q7_4 0.85 0.85 0.84 0.44 5.5 0.013 0.0113 0.45
Q7_5 0.83 0.83 0.83 0.42 5.0 0.014 0.0124 0.42
Q7_7 0.83 0.84 0.84 0.42 5.1 0.014 0.0164 0.41
Q7_8 0.84 0.84 0.84 0.43 5.3 0.013 0.0146 0.45
Q7_12 0.85 0.85 0.85 0.44 5.6 0.013 0.0188 0.45
Q7_13 0.87 0.87 0.86 0.48 6.5 0.011 0.0083 0.47
Q7_14 0.85 0.85 0.85 0.45 5.7 0.013 0.0185 0.47
Item statistics
n raw.r std.r r.cor r.drop mean sd
Q7_2 328 0.70 0.71 0.66 0.61 2.7 0.86
Q7_4 328 0.71 0.71 0.68 0.60 2.8 0.98
Q7_5 328 0.80 0.80 0.79 0.72 2.9 0.95
Q7_7 328 0.79 0.78 0.75 0.70 3.7 1.01
Q7_8 328 0.74 0.75 0.70 0.65 2.8 0.88
Q7_12 328 0.72 0.71 0.65 0.61 3.4 1.02
Q7_13 328 0.57 0.56 0.47 0.43 3.6 0.99
Q7_14 328 0.69 0.69 0.62 0.58 3.6 0.99
Non missing response frequency for each item
1 2 3 4 5 miss
Q7_2 0.11 0.21 0.57 0.09 0.02 0
Q7_4 0.10 0.21 0.47 0.17 0.05 0
Q7_5 0.10 0.18 0.51 0.17 0.04 0
Q7_7 0.05 0.09 0.17 0.53 0.16 0
Q7_8 0.10 0.17 0.55 0.15 0.02 0
Q7_12 0.07 0.09 0.32 0.43 0.10 0
Q7_13 0.02 0.11 0.32 0.36 0.19 0
Q7_14 0.05 0.08 0.24 0.48 0.15 0
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] xtable_1.8-4 kableExtra_1.3.1 readxl_1.3.1 coda_0.19-4
[5] nFactors_2.4.1 lattice_0.20-41 psych_2.0.12 psychometric_2.2
[9] multilevel_2.6 MASS_7.3-53 nlme_3.1-151 mvtnorm_1.1-1
[13] ggcorrplot_0.1.3 naniar_0.6.0 simsem_0.5-15 lslx_0.6.10
[17] MIIVsem_0.5.5 lavaanPlot_0.5.1 semTools_0.5-4 lavaan_0.6-7
[21] data.table_1.13.6 patchwork_1.1.1 forcats_0.5.0 stringr_1.4.0
[25] dplyr_1.0.3 purrr_0.3.4 readr_1.4.0 tidyr_1.1.2
[29] tibble_3.0.5 ggplot2_3.3.3 tidyverse_1.3.0
loaded via a namespace (and not attached):
[1] fs_1.5.0 lubridate_1.7.9.2 webshot_0.5.2 RColorBrewer_1.1-2
[5] httr_1.4.2 rprojroot_2.0.2 tools_4.0.3 backports_1.2.0
[9] R6_2.5.0 DBI_1.1.1 colorspace_2.0-0 withr_2.4.0
[13] tidyselect_1.1.0 mnormt_2.0.2 compiler_4.0.3 git2r_0.28.0
[17] cli_2.2.0 rvest_0.3.6 xml2_1.3.2 scales_1.1.1
[21] digest_0.6.27 pbivnorm_0.6.0 rmarkdown_2.6 pkgconfig_2.0.3
[25] htmltools_0.5.1 highr_0.8 dbplyr_2.0.0 htmlwidgets_1.5.3
[29] rlang_0.4.10 rstudioapi_0.13 visNetwork_2.0.9 generics_0.1.0
[33] jsonlite_1.7.2 magrittr_2.0.1 Rcpp_1.0.6 munsell_0.5.0
[37] fansi_0.4.2 lifecycle_0.2.0 visdat_0.5.3 stringi_1.5.3
[41] whisker_0.4 yaml_2.2.1 grid_4.0.3 parallel_4.0.3
[45] promises_1.1.1 crayon_1.3.4 haven_2.3.1 hms_1.0.0
[49] tmvnsim_1.0-2 knitr_1.30 ps_1.5.0 pillar_1.4.7
[53] stats4_4.0.3 reprex_0.3.0 glue_1.4.2 evaluate_0.14
[57] modelr_0.1.8 vctrs_0.3.6 httpuv_1.5.5 cellranger_1.1.0
[61] gtable_0.3.0 assertthat_0.2.1 xfun_0.20 broom_0.7.3
[65] later_1.1.0.1 viridisLite_0.3.0 workflowr_1.6.2 DiagrammeR_1.0.6.1
[69] ellipsis_0.3.1