Sample Size Determination
Last updated: 2021-02-18
Checks: 7 0
Knit directory: pools-projects/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201007) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ea0a6a6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/development-study-CFA-invariance-test.Rmd
Untracked: analysis/development-study-CFA.Rmd
Untracked: analysis/development-study-EFA.Rmd
Untracked: analysis/development-study-data-management.Rmd
Untracked: analysis/study-1-power-calculation.Rmd
Untracked: code/laplace_functions.R
Untracked: code/pdf2png.R
Untracked: code/utility_functions.R
Untracked: data/efa_results_2021_01_06.csv
Untracked: data/fit-test.RData
Untracked: data/savedlocalfit.RData
Untracked: diagrams/
Untracked: item-review-2/expert-review-2-response1.pdf
Untracked: item-review-2/expert-review-2-response2.pdf
Untracked: item-review-2/expert-review-2-response3.pdf
Untracked: item-review-2/pilot-data-item-review.xlsx
Untracked: manuscript/
Untracked: output/cfa-final-parameterEstimates.csv
Untracked: output/cfa_results.csv
Untracked: output/corr-plot.pdf
Untracked: output/corr-residuals.pdf
Unstaged changes:
Modified: .Rprofile
Deleted: .gitattributes
Modified: .gitignore
Modified: analysis/index.Rmd
Deleted: analysis/pilot-study-CFA.Rmd
Deleted: analysis/pilot-study-EFA.Rmd
Deleted: analysis/pilot-study-data-management.Rmd
Deleted: analysis/pilot-study-power-calculation.Rmd
Modified: code/load_packages.R
Modified: item-review-1/response8_nov6.pdf
Modified: item-review-2/Overview of Expert Review v2.0 Results.docx
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/power-calculation.Rmd) and HTML (docs/power-calculation.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 4856250 | noah-padgett | 2020-11-19 | pilot-efa-results |
| html | 4856250 | noah-padgett | 2020-11-19 | pilot-efa-results |
POOLS Model
The POOLS model is generally a four factor model with 10-14 items per factor.
Power Analysis Simulation Study
Here, we vary the sample size between 100 and 1000 (steps of 50) to determine the approximate minimal sample size needed to reject that the data fails to fit the model.
Populaiton Model
pop_model <- "
# POOLS Items (~40)
EF =~ .6*p11 + .6*p12 + .6*p13 + .6*p14 + .6*p15 + .6*p16 + .6*p17 + .6*p18 + .6*p19 + .6*p110
ST =~ .6*p21 + .6*p22 + .6*p23 + .6*p24 + .6*p25 + .6*p26 + .6*p27 + .6*p28 + .6*p29 + .6*p210
IN =~ .6*p31 + .6*p32 + .6*p33 + .6*p34 + .6*p35 + .6*p36 + .6*p37 + .6*p38 + .6*p39 + .6*p310
EN =~ .6*p41 + .6*p42 + .6*p43 + .6*p44 + .6*p45 + .6*p46 + .6*p47 + .6*p48 + .6*p49 + .6*p410
# Self-Efficacy Items (12)
# omegas: .85, .82, .8
se1 =~ .75*eff1 + .75*eff2 + .75*eff3 +.75*eff4
se2 =~ .75*eff5 + .75*eff6 + .7*eff7 +.7*eff8
se3 =~ .7*eff9 + .7*eff10 + .7*eff11 +.7*eff12
# Teacher-Team Inno Scale (omega = .89)
tt =~ .8*ttis1 + .8*ttis2 + .8*ttis3 + .8*ttis4
# NEO FFM
# Alphas: .77, .81, .82, .86, .86
A =~ .7*ffmA1 + .7*ffmA2 + .7*ffmA3 + .7*ffmA4 + .7*ffmA5 + .7*ffmA6 + .7*ffmA7 + .7*ffmA8 + .7*ffmA9 + .7*ffmA10
C =~ .7*ffmC1 + .7*ffmC2 + .7*ffmC3 + .7*ffmC4 + .7*ffmC5 + .7*ffmC6 + .7*ffmC7 + .7*ffmC8 + .7*ffmC9 + .7*ffmC10
O =~ .7*ffmO1 + .7*ffmO2 + .7*ffmO3 + .7*ffmO4 + .7*ffmO5 + .7*ffmO6 + .7*ffmO7 + .7*ffmO8 + .7*ffmO9 + .7*ffmO10
E =~ .7*ffmE1 + .7*ffmE2 + .7*ffmE3 + .7*ffmE4 + .7*ffmE5 + .7*ffmE6 + .7*ffmE7 + .7*ffmE8 + .7*ffmE9 + .7*ffmE10
N =~ .7*ffmN1 + .7*ffmN2 + .7*ffmN3 + .7*ffmN4 + .7*ffmN5 + .7*ffmN6 + .7*ffmN7 + .7*ffmN8 + .7*ffmN9 + .7*ffmN10
# Latent Variable Covariance Matrix
# POOLS
EF ~~ 1*EF + .3*ST + .2*In + .2*EN
ST ~~ 1*ST + .1*IN + .3*EN
IN ~~ 1*IN + .3*EN
EN ~~ 1*EN
# SE
se1 ~~ 1*se1 + .5*se2 + .4*se3
se2 ~~ 1*se2 + .3*se3
se3 ~~ 1*se3
# FFM (taken from deyoung table 6)
A ~~ 1*A + (0.38)*C + (0.11)*O + (0.15)*E + (-0.24)*N
C ~~ 1*C + (0.11)*O + (0.18)*E + (-0.24)*N
O ~~ 1*O + (0.26)*E + (-0.13)*N
E ~~ 1*E + (-0.33)*N
N ~~ 1*N
# Research questions
# 1. FFM accounting for variance of POOLS
# (need to come up with better values)
EF ~ .1*A + .3*C + .4*O + .1*E + (-0.2)*N
ST ~ .1*A + .3*C + .4*O + .1*E + (-0.2)*N
EN ~ .1*A + .3*C + .4*O + .1*E + (-0.2)*N
IN ~ .1*A + .3*C + .4*O + .1*E + (-0.2)*N
# 2. relationship between SE & Pools
EF ~ .3*se1 + .2*se2 + .4*se3
ST ~ .3*se1 + .2*se2 + .4*se3
IN ~ .3*se1 + .2*se2 + .4*se3
EN ~ .3*se1 + .2*se2 + .4*se3
# 3. Team inno predicting POOLS
EF ~ .2*tt
ST ~ .2*tt
IN ~ .2*tt
EN ~ .2*tt
# 4. demographics
EF ~ 0.1*sex
ST ~ 0.1*sex
IN ~ 0.1*sex
EN ~ 0.1*sex
# categorical variables
sex | 0.4*t1
"
dat <- simulateData(pop_model, model.type = "sem")Warning in lav_partable_flat(FLAT, blocks = "group", meanstructure =
meanstructure, : lavaan WARNING: thresholds are defined for exogenous variables:
sex#Impose missing
datmiss <- imposeMissing(
dat,
nforms = 7,
itemGroups = list(c(1:107),
c(),
c(41:56),
c( 77:86, 87:96, 97:106),
c( 57:66, 87:96, 97:106),
c( 57:66, 67:76, 97:106),
c( 57:66, 67:76, 77:86 ),
c( 67:76, 77:86, 87:96 )
))
naniar::vis_miss(datmiss)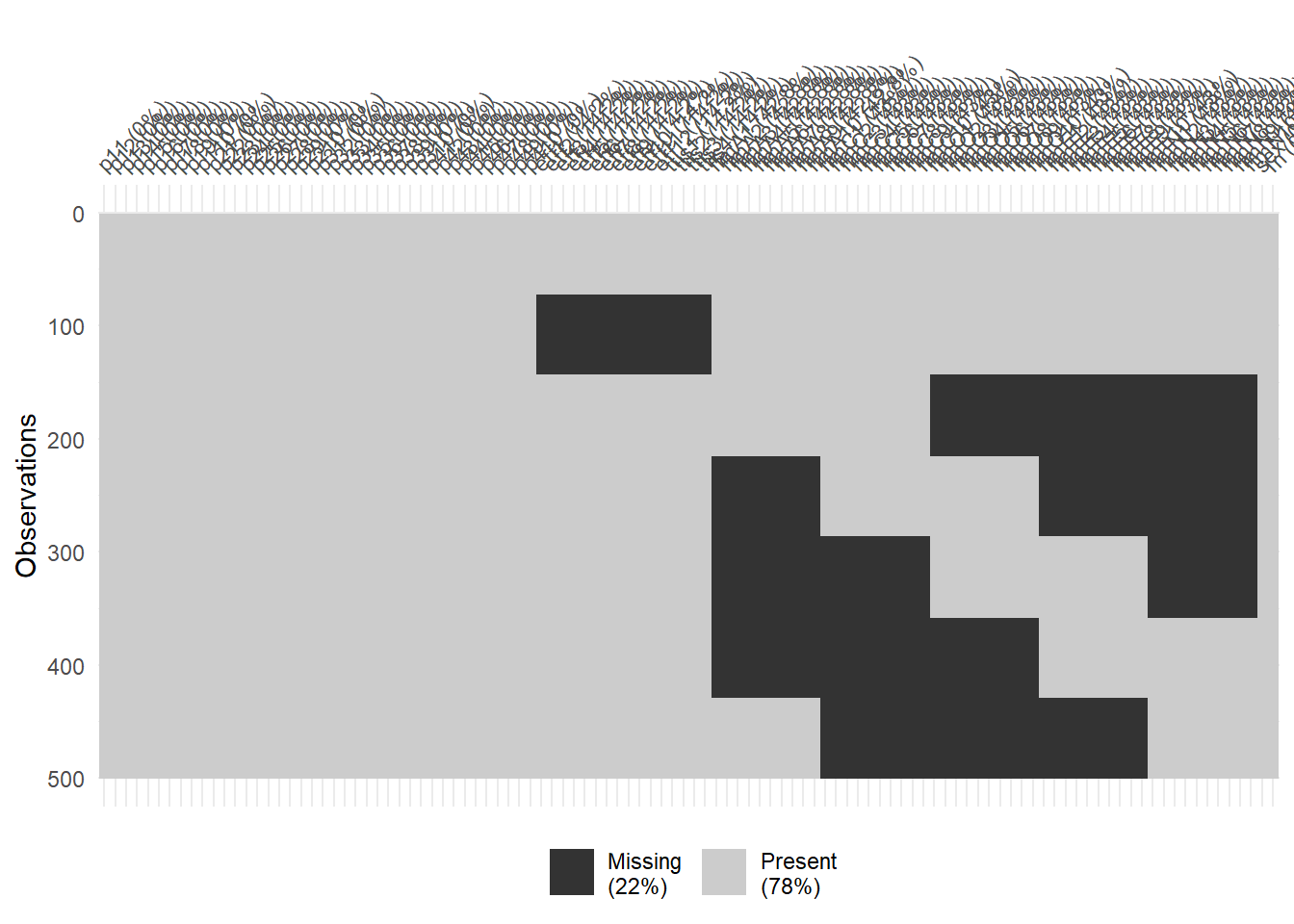
Estimation Model
est_model <- '
# 1. Latent variable definition
# POOLS
EF =~ 1*p11 + p12 + p13 + p14 + p15# + p16 + p17 + p18 + p19 + p110
ST =~ 1*p21 + p22 + p23 + p24 + p25# + p26 + p27 + p28 + p29 + p210
IN =~ 1*p31 + p32 + p33 + p34 + p35# + p36 + p37 + p38 + p39 + p310
EN =~ 1*p41 + p42 + p43 + p44 + p45# + p46 + p47 + p48 + p49 + p410
# Self-Efficacy Items
se1 =~ NA*eff1 + eff2 + eff3 + eff4
se2 =~ NA*eff5 + eff6 + eff7 + eff8
se3 =~ NA*eff9 + eff10 + eff11 + eff12
# Teacher-Team Inno Scale
tt =~ NA*ttis1 + ttis2 + ttis3 + ttis4
# NEO FFM
A =~ NA*ffmA1 + ffmA2 + ffmA3 + ffmA4 + ffmA5 + ffmA6 + ffmA7 + ffmA8 + ffmA9 + ffmA10
C =~ NA*ffmC1 + ffmC2 + ffmC3 + ffmC4 + ffmC5 + ffmC6 + ffmC7 + ffmC8 + ffmC9 + ffmC10
O =~ NA*ffmO1 + ffmO2 + ffmO3 + ffmO4 + ffmO5 + ffmO6 + ffmO7 + ffmO8 + ffmO9 + ffmO10
E =~ NA*ffmE1 + ffmE2 + ffmE3 + ffmE4 + ffmE5 + ffmE6 + ffmE7 + ffmE8 + ffmE9 + ffmE10
N =~ NA*ffmN1 + ffmN2 + ffmN3 + ffmN4 + ffmN5 + ffmN6 + ffmN7 + ffmN8 + ffmN9 + ffmN10
# 2. Latent variable covariances
# POOLS
EF ~~ EF + ST + IN + EN
ST ~~ ST + IN + EN
IN ~~ IN + EN
# SE
se1 ~~ 1*se1 + se2 + se3
se2 ~~ 1*se2 + se3
se3 ~~ 1*se3
# Team Inno.
tt ~~ 1*tt
# FFM
A ~~ 1*A + C + O + E + N
C ~~ 1*C + O + E + N
O ~~ 1*O + E + N
E ~~ 1*E + N
N ~~ 1*N
# Research questions
# 1. FFM accounting for variance of POOLS
EF ~ A + C + O + E + N
ST ~ A + C + O + E + N
IN ~ A + C + O + E + N
EN ~ A + C + O + E + N
# 2. relationship between SE & Pools
EF ~ se1 + se2 + se3
ST ~ se1 + se2 + se3
IN ~ se1 + se2 + se3
EN ~ se1 + se2 + se3
# 3. Team inno predicting POOLS
EF ~ tt
ST ~ tt
IN ~ tt
EN ~ tt
'
# use missing = "ML" for FIML
fit <- cfa(est_model, datmiss, estimator = "ML", missing = "ML")
summary(fit, standardized=T, fit.measures=T)
# ================================== #
# funciton:
# data_function_categorization()
#
# Purpose:
# categorize the continuous response
# into the 5 discrete categories we
# will observe in the analysis.
# We plan to treat the data as continuous.
# But, we will also use a robust estimation
# method DWLS and
# PML (pairwise maximum likelihood)
data_function_categorization <- function(data){
tauCreate <- function(x){
e <- rnorm(4,0, 0.01)
BREAKS <- c(-Inf, -1.4+e[1], -0.4+e[2], 0.2+e[3], 1+e[4], Inf)
x <- cut(x,
breaks=BREAKS,
labels = c(-2, -1, 0, 1, 2))
as.numeric(x)-3 # center at 0
}
data[,1:106] <- apply(data[,1:106], 2, tauCreate)
data
}
missdata_mech <- miss(
nforms = 7,
itemGroups = list(c(1:107),
c(),
c(41:56),
c( 77:86, 87:96, 97:106),
c( 57:66, 87:96, 97:106),
c( 57:66, 67:76, 97:106),
c( 57:66, 67:76, 77:86 ),
c( 67:76, 77:86, 87:96 ))
)
sim_res <- sim(
nRep = 5, n = 500,
lavaanfun = "sem",
model = list(model=est_model, estimator = "ML", missing = "ML"),
generate = pop_model,
miss = missdata_mech,
datafun = data_function_categorization
)
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] xtable_1.8-4 kableExtra_1.3.1 readxl_1.3.1 coda_0.19-4
[5] nFactors_2.4.1 lattice_0.20-41 psych_2.0.12 psychometric_2.2
[9] multilevel_2.6 MASS_7.3-53 nlme_3.1-151 mvtnorm_1.1-1
[13] ggcorrplot_0.1.3 naniar_0.6.0 simsem_0.5-15 lslx_0.6.10
[17] MIIVsem_0.5.5 lavaanPlot_0.5.1 semTools_0.5-4 lavaan_0.6-7
[21] data.table_1.13.6 patchwork_1.1.1 forcats_0.5.0 stringr_1.4.0
[25] dplyr_1.0.3 purrr_0.3.4 readr_1.4.0 tidyr_1.1.2
[29] tibble_3.0.5 ggplot2_3.3.3 tidyverse_1.3.0
loaded via a namespace (and not attached):
[1] fs_1.5.0 lubridate_1.7.9.2 webshot_0.5.2 RColorBrewer_1.1-2
[5] httr_1.4.2 rprojroot_2.0.2 tools_4.0.3 backports_1.2.0
[9] R6_2.5.0 DBI_1.1.1 colorspace_2.0-0 withr_2.4.0
[13] tidyselect_1.1.0 mnormt_2.0.2 compiler_4.0.3 git2r_0.28.0
[17] cli_2.2.0 rvest_0.3.6 xml2_1.3.2 labeling_0.4.2
[21] scales_1.1.1 digest_0.6.27 pbivnorm_0.6.0 rmarkdown_2.6
[25] pkgconfig_2.0.3 htmltools_0.5.1 dbplyr_2.0.0 htmlwidgets_1.5.3
[29] rlang_0.4.10 rstudioapi_0.13 farver_2.0.3 visNetwork_2.0.9
[33] generics_0.1.0 jsonlite_1.7.2 magrittr_2.0.1 Rcpp_1.0.6
[37] munsell_0.5.0 fansi_0.4.2 lifecycle_0.2.0 visdat_0.5.3
[41] stringi_1.5.3 whisker_0.4 yaml_2.2.1 grid_4.0.3
[45] parallel_4.0.3 promises_1.1.1 crayon_1.3.4 haven_2.3.1
[49] hms_1.0.0 tmvnsim_1.0-2 knitr_1.30 ps_1.5.0
[53] pillar_1.4.7 stats4_4.0.3 reprex_0.3.0 glue_1.4.2
[57] evaluate_0.14 modelr_0.1.8 vctrs_0.3.6 httpuv_1.5.5
[61] cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1 xfun_0.20
[65] broom_0.7.3 later_1.1.0.1 viridisLite_0.3.0 workflowr_1.6.2
[69] DiagrammeR_1.0.6.1 ellipsis_0.3.1